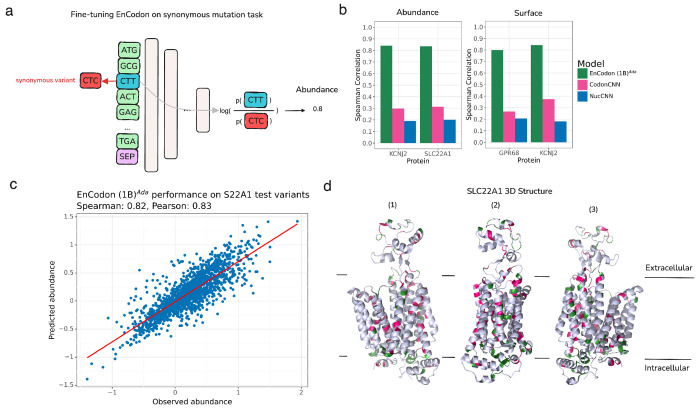
Figure 5: EnCodon (1B)Ada generalizes well across unseen synonymous variants in membrane proteins.
a) We re-purposed the pre-trained language modeling classifier head for synonymous mutation effect modeling. Specifically, given a synonymous codon variant, we first compute the codon likelihoods (i.e. logits) for the variant’s position. Next, the log-ratio of wild-type codon against mutated codon is considered as the final variant’s effect prediction – protein abundance level or surface expression in this experiment. Notably, using no additional weights, the mutation’s effect on protein’s abundance measurement is modeled as the log-likelihood ratio between mutated and wild-type codon given the wild-type coding sequence in input. b) Spearman correlation between predicted and observed abundance (left bar plot) or surface expression (right bar plot) were shown for test synonymous variants in KCNJ2, SLC22A1, and GPR68 proteins. c) An test set of synonymous variants applied on SLC22A1 were held-out from the training data of the EnCodons. The scatter plot of predicted vs. observed abundance is shown for eukaryotic adapted EnCodon (1B) which showed as the top-performed compared to other fine-tuned EnCodons. d) After fine-tuning, we performed in-silico synonymous mutagenesis with the best-performing EnCodon model. We selected “critical” synonymous variants for which the predicted abundance was above 95-th (green) or below the 5-th quantile (pink). Next, extracted SLC22A1 extreme variants were overlayed in the protein’s 3D structure which is shown from 3 different angles.