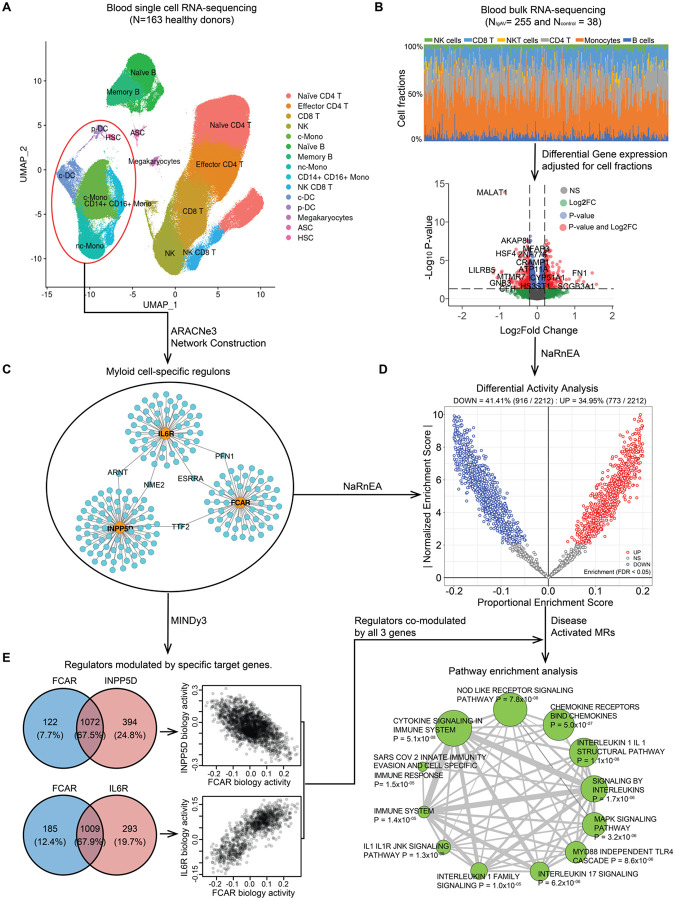
Figure 5. Master regulators of IgA vasculitis modulated by the GWAS candidate genes.
(a) the UMAP plot for the reference single cell RNA-seq data from 163 healthy individuals. The blood cells were clustered and annotated into 15 different immune cell types highlighted in different colors; (b) Deconvolution and differential gene expression analysis based on blood bulk RNA-seq data from 255 IgAV cases and 38 health controls. The upper panel shows the distribution of different cell type fractions across all 293 individuals. The volcano plot (lower panel: x-axis denotes the log2 Fold Change and the y-axis indicates the adjusted P values) with differentially expressed genes in red (Padjusted<0.05 and log2 Fold Change>0.2) after adjusting for cell type fractions; (c) The myeloid regulons reconstructed transcriptome-wide based on the myeloid lineage cells (circled in red above) using ARACNe3 software (a subset of only 3 regulons shown to illustrate this step). (d) Differential activity analysis based on the myeloid regulons and cell fraction-adjusted differential gene expression signature in IgAV cases compared to controls. The x-axis indicates the proportional enrichment score (effect size for gene set enrichment that was normalized by set’s size, parameterization, and gene expression signature). The y-axis shows the normalized score compared to the expectation under the null hypothesis. The red and blue colors indicate the regulators with higher and lower activity, respectively, in IgAV cases compared to healthy controls. (e) Regulators co-modulated by the GWAS candidate genes: 1,072 regulators co-modulated by FCAR and INPP5D with mostly opposed effects on their biological activities (upper panel) and 1,009 regulators co-modulated by FCAR and IL6R with direction consistent effects on their biological activities (lower panel). The targets of the top master regulators co-modulated by all 3 genes were enriched in multiple immune related pathways based on FDR< 0.05 (depicted as enrichment map with a node size proportional to the statistical significance of enrichment and edge thickness proportional to gene set overlaps).