Figure 3.
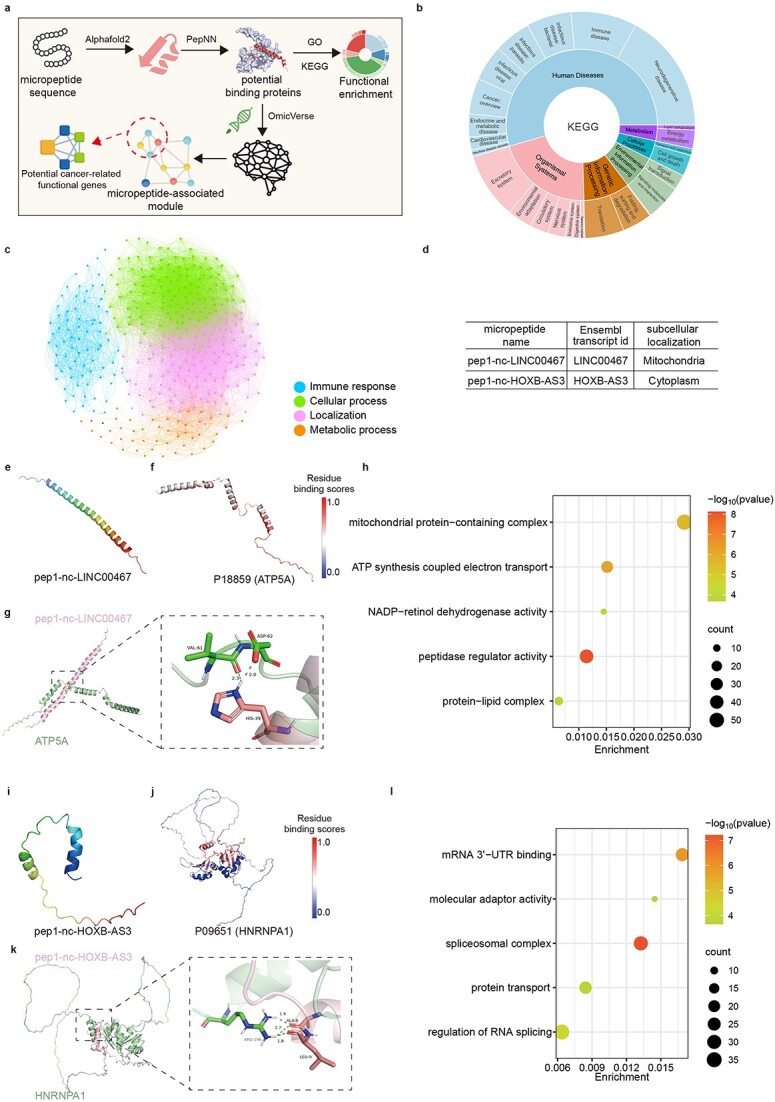
Exploration of micropeptide structures and biological functions. (a) Overview of the micropeptide functional annotation design pipeline. (b) Comprehensive visualization of KEGG pathways by Plotly-sunburst method. (c) GO classification atlas for micropeptides based on potential protein interactions. Candidates are categorized into four groups. (d) Two previous research of micropeptide information in HMPA. (e) The 3D structure of pep1-nc-LINC00467 predicted by AlphaFold2. (f) Visualization ATP5A each residue binding probability colored by B-factor, range for probability is [0,1]. (g) Docking result of pep1-nc-LINC00467 and P18859 by ClusPro. (h) The 3D structure of pep1-nc-HOXB-AS3 predicted by AlphaFold2. (i) Visualization HNRNRA1 each residue binding probability colored by B-factor, range for probability is [0,1]. (j) Docking result of pep1-nc-HOXB-AS3 and P09651 by ClusPro. (k) GO enrichment result for proteins in mPPI network. (l) GO enrichment result for proteins in mPPI interaction network.
