Table 3.
Quantitative results of various model variants evaluated on STARmap and MERFISH datasets, focusing on performance metrics including PCC, SPCC, SSIM, RMSE, and COSSIM. These metrics provide a comprehensive assessment of the models’ ability to accurately reconstruct spatial gene expression patterns.
| Variants | STARmap | MERFISH | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
PCC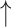
|
SPCC 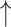
|
SSIM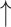
|
RMSE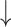
|
COSSIM
|
PCC
|
SPCC 
|
SSIM
|
RMSE
|
COSSIM
|
|
| Module-Variant-1 | 0.2099 | 0.1845 | 0.0842 | 1.2722 | 0.4015 | 0.3125 | 0.2514 | 0.2015 | 1.1615 | 0.4215 |
| Module-Variant-2 | 0.2155 | 0.1884 | 0.0812 | 1.2451 | 0.4154 | 0.3348 | 0.2678 | 0.2248 | 1.1443 | 0.4412 |
| Loss-Variant-1 | 0.1654 | 0.1654 | 0.0611 | 1.2978 | 0.3889 | 0.2484 | 0.1814 | 0.1872 | 1.2311 | 0.3548 |
| Loss-Variant-2 | 0.1933 | 0.1789 | 0.0074 | 1.2845 | 0.3945 | 0.2778 | 0.2019 | 0.0187 | 1.2102 | 0.3784 |
| SELF-Former | 0.2359 | 0.2330 | 0.1016 | 1.2327 | 0.4387 | 0.3566 | 0.2780 | 0.2493 | 1.1270 | 0.4692 |
