Fig. 4. Fluorescence and gel-based readout of the RADD assay.
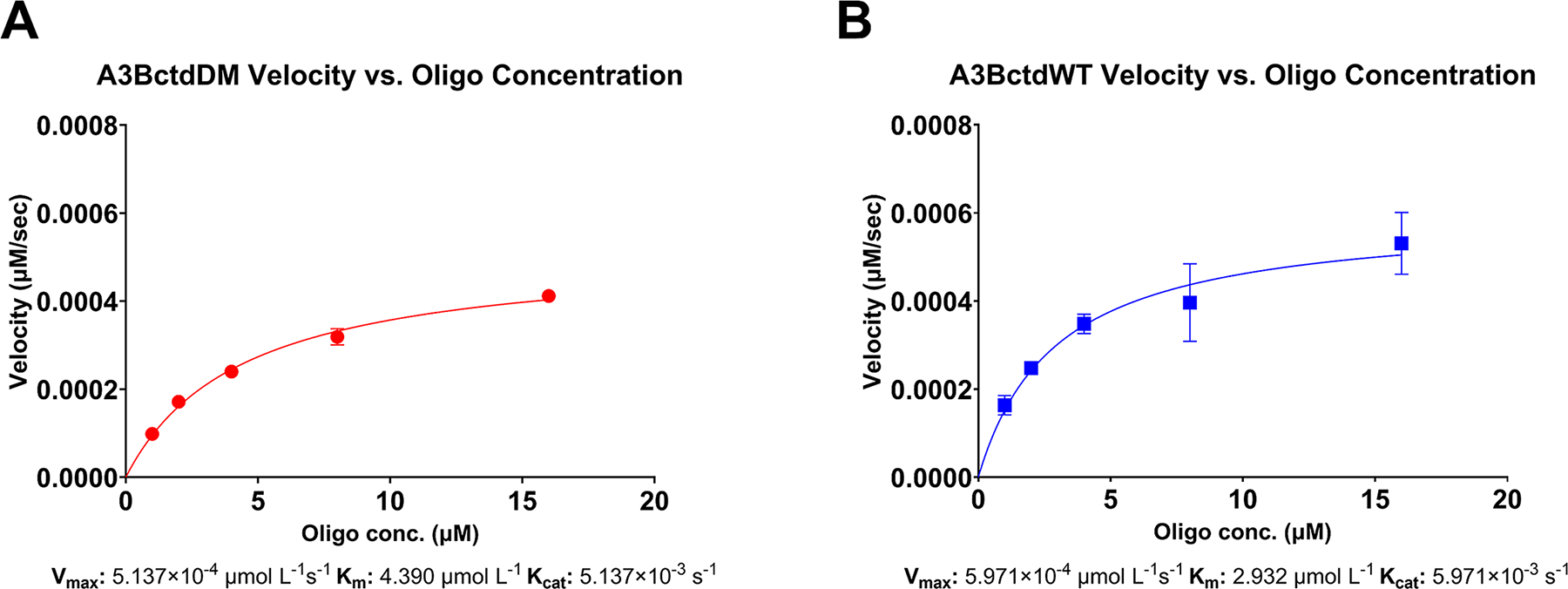
(A) Enzyme velocity of 100 nM A3BctdDM versus a ½ dilution series of 16 μM down to 1 μM reporter oligo (N = 3) fitted to the Michaelis-Menten model (R2 = 0.99). The predicted Vmax was 5.1397 × 10−4 μmol/L/s 95% CI [4.789 × 10−4–5.532 × 10−4], resulting in a Kcat of 5.137 × 10−3 s−1. The Km of A3BctdDM was predicted to be 4.390 μmol/L 95% CI [3.651–5.290]. (B) Enzyme velocity of A3BctdWT versus the same reporter oligo dilution series described above (N = 3), fitted to the Michaelis-Menten model (R2 = 0.87). The predicted Vmax for A3BctdWT was 5.971 × 10−4 μmol/L/s 95 % CI [5.051 × 10−4, 7.204 × 10−4], resulting in a Kcat of 5.971 × 10−3/s. The Km was predicted to be 2.932 μmol/L 95% CI [1.723, 4.943].
Figure adapted from Belica, C., Carpenter, M. A., Chen, Y., Moeller, N., Harris, R. S., & Aihara, H. (2024). A real-time biochemical assay for quantitative analyses of APOBEC-catalyzed DNA deamination. Journal of Biological Chemistry.
