Fig. 5. Estimating first-order rate constants using RADD and sequential enzymatic reaction modeling.
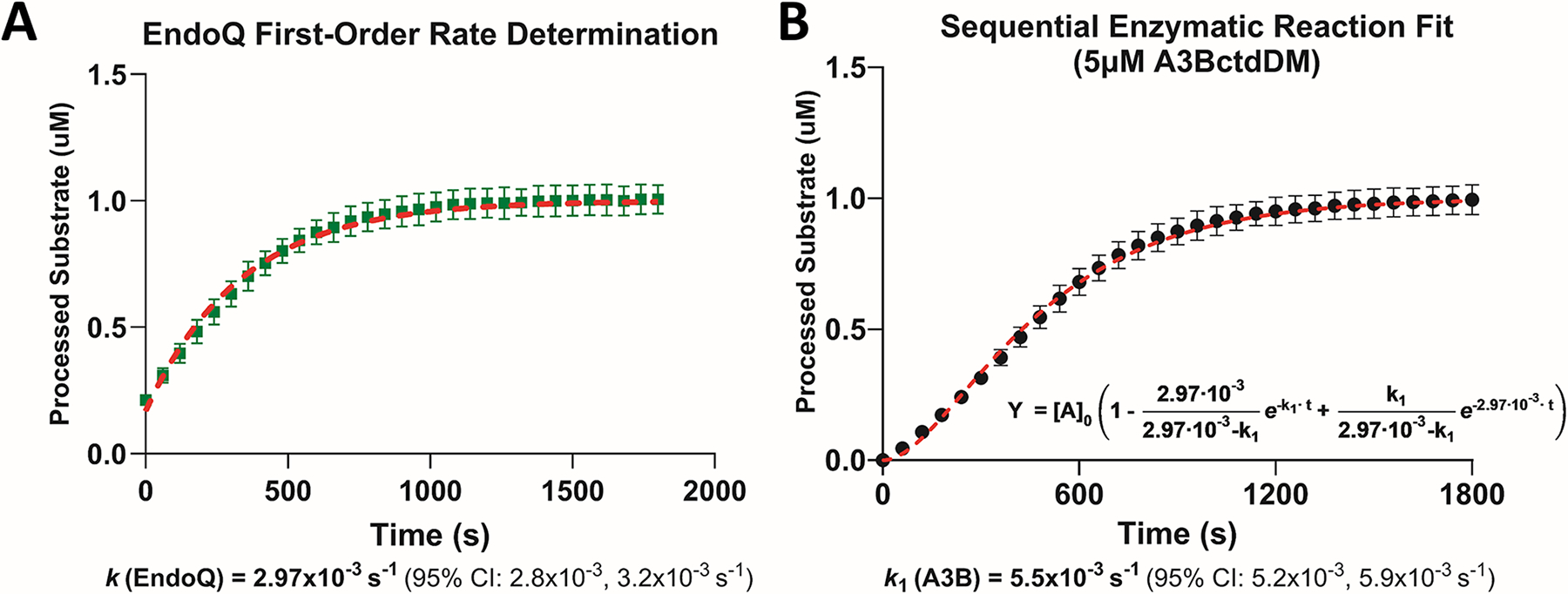
(A) Estimating the kcat of EndoQ by measuring the kobs of EndoQ in a single-turnover condition (N = 3). The kcat of EndoQ was determined to be 2.97 × 10−3/s 95% CI [2.8 × 10−3–3.2 × 10−3] (R2 = 0.97). (B) Estimating the kcat of A3BctdDM by measuring the deamination kobs at 5 μM A3BctdDM (N = 3). Using the kcat of EndoQ as k2 in the sequential enzyme reaction equation, the model showed an excellent fit to the A3BctdDM data (R2 = 0.98) and predicted a kcat of 5.5 × 10−3/s 95% CI [5.2 × 10−3–5.9 × 10−3/s].
Figure adapted from Belica, C., Carpenter, M. A., Chen, Y., Moeller, N., Harris, R. S., & Aihara, H. (2024). A real-time biochemical assay for quantitative analyses of APOBEC-catalyzed DNA deamination. Journal of Biological Chemistry.
