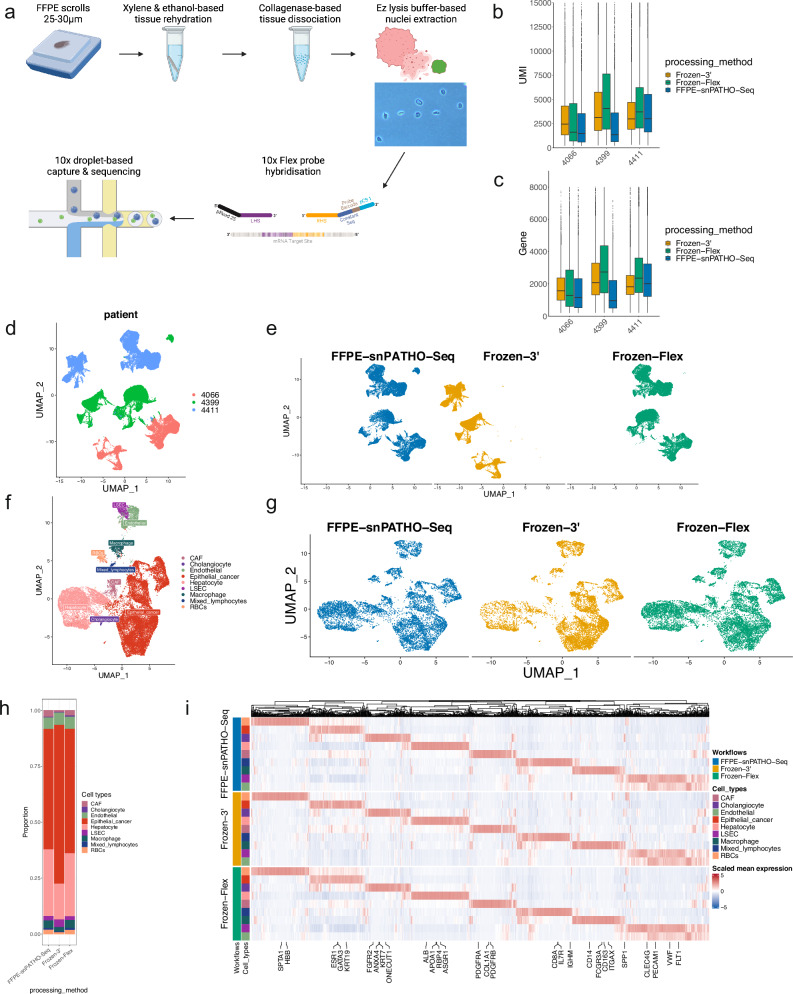
Fig. 2. The snPATHO-seq workflow enables nuclei isolation and single-nucleus gene expression detection from human FFPE tissue samples.
a Illustration of the snPATHO-seq workflow. Created in BioRender. Wang, T. (2024) BioRender.com/u53s150. b, c Boxplots of the number of UMIs (b) and genes (c) detected per nucleus. The boxes show the UMIs (b) and Genes (c) median and interquartile range. Outliers were shown as dots. d UMAP embedding of unintegrated snRNA-seq data annotated by sample IDs. e UMAP embedding of unintegrated snRNA-seq data split by processing methods. f UMAP embedding of Seurat CCA integrated snRNA-seq data from patient 4411 annotated by cell type. g UMAP embedding of Seruat CCA integrated 4411 snRNA-seq data split by processing methods. h Barplot showing the fraction of cell types detected by different snRNA-seq methods in sample 4411. i Heatmap of the scaled expression of selected cell type markers detected by differential gene expression analyses in 4411 data. The top 200 significantly differentially expressed genes identified in each cell population (if available) were selected by fold change and used for plotting. A gene was considered significantly differentially expressed if the BH-adjusted P value was lower than 0.05. Genes were arranged by hierarchical clustering based on the expression in the FFPE-snPATHO-seq data on the x-axis. Cell types identified by different snRNA-seq workflows were manually arranged on the y-axis. N = 1 sample per protocol.