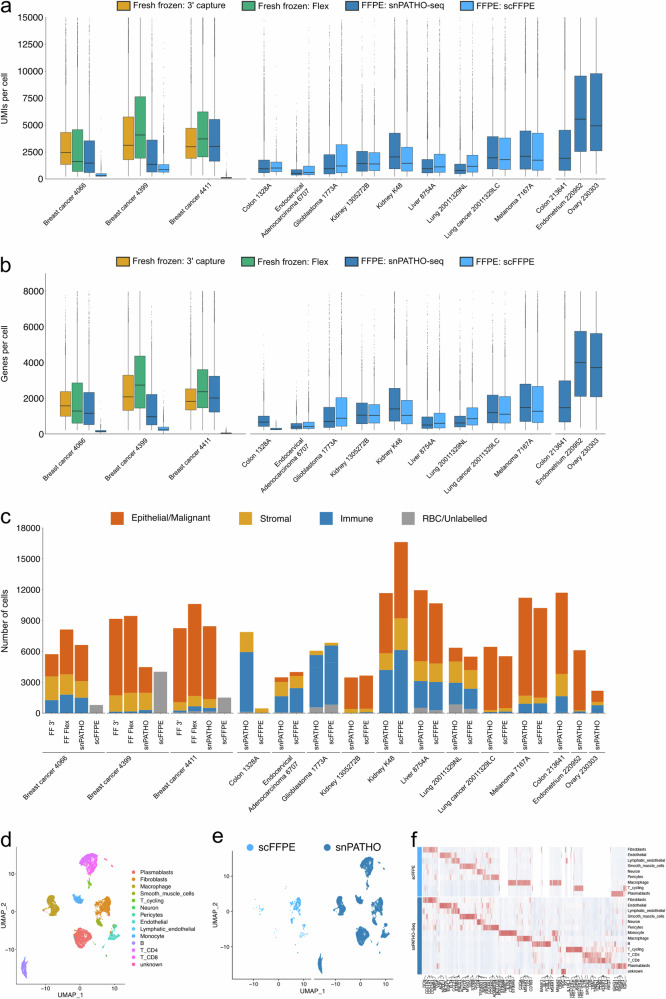
Fig. 4. Comparison of the snPATHO-seq workflow to scFFPE workflow.
a, b Boxplots of the number of UMIs (a) and genes (b) detected per nuclei using different workflows. The boxes show the median and interquartile range of the UMIs (a) and Genes (b). Outliers were shown as dots. c Barplots of the number of nuclei/cells detected in each dataset colored by major cell lineages. d UMAP embedding of Seurat CCA integrated snPATHO-seq and scFFPE data from sample Colon_1328A colored by cell type annotations. e UMAP embedding split by processing methods. f Heatmap of top differentially expressed genes detected between cell types by snPATHO-seq and scFFPE workflows. The top 200 significantly differentially expressed genes identified in each cell population (if available) were selected by fold change and used for plotting. A gene was considered significantly differentially expressed if the BH-adjusted P value was lower than 0.05. N = 1 sample per protocol.