Figure 6.
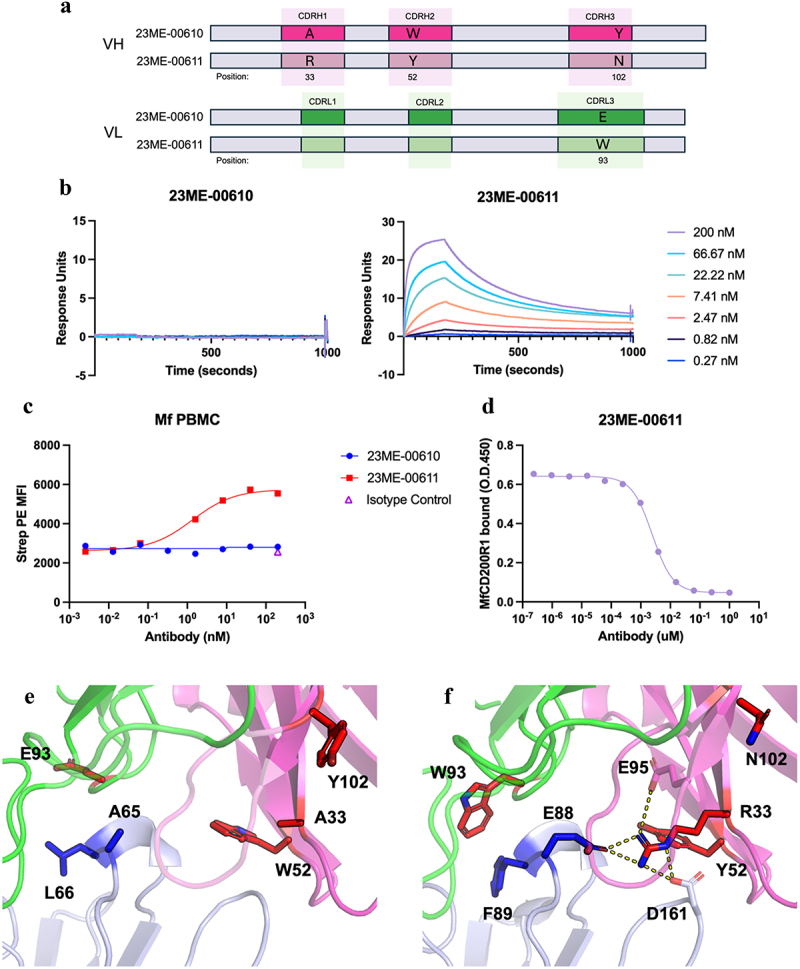
Mutations in 23ME-00611 surrogate antibody confers high affinity binding to MfCD200R1. (a) Schematics that show the VH or VL CDR positions mutated from 23ME-00610 to 23ME-00611; (b) biacore sensorgrams that show lack of 23ME-00610 binding to MfCD200R1 with concentration up to 200 nM (left) and 23ME-00611’s binding to MfCD200R1 at KD = 2 nM at 37°C (right); (c) cynomolgus PBMC FACS study confirms lack of 23ME-00610 binding to MfCD200R1 while 23ME-00611showed good binding potency. (d) 23ME-00611’s potent inhibition of MfCD200R1:MfCD200 interaction as shown by ELISA; (e) zoom-in views of 23ME-00610:hCD200R1 crystal structure and (f) 23ME-00611:MfCD200R1AlphaFold model show the two structurally equivalent residues (blue sticks) in hCD200R1 or MfCD200R1 that can explain the lack of 23ME-0610 binding and the four fab mutations in red sticks that confer 23ME-00611 high affinity binding to MfCD200R1. 23ME-00611 A33R mutation permits a network of charge interaction (dotted line) to accommodate E88 in MfCD200R1. Heavy and light chains indexed using kabat and chothia numbering, respectively.22–24
