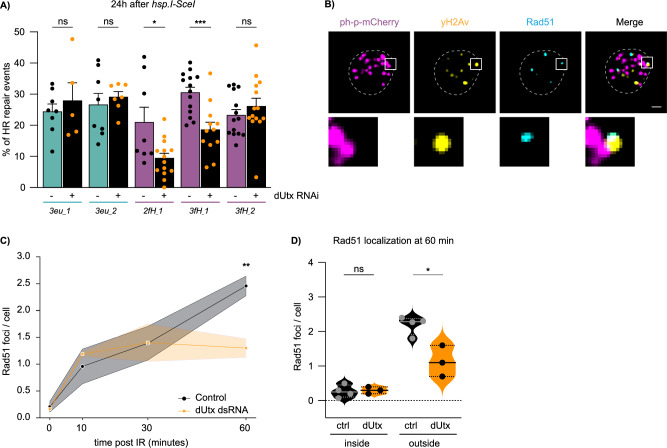
Fig. 5. dUtx is required for homologous recombination in facultative heterochromatin.
A DR-white/hsp.I-SceI larvae with (+) or without (˗) dUtx RNAi were heat-shocked for one hour to induce I-SceI. Repair PCR products were Sanger sequenced 24 h after I-SceI induction and analyzed using the TIDE algorithm37. Graph shows percentage of identified homologous recombination (HR) products. Bars indicate averages +SEM of n = 14 single larvae (biological replicates) per condition (except 3eu_1, 3eu_2, 2fH_1: n = 8, 3eu_1 (dUtx RNAi): n = 5, 3eu_2 (dUtx RNAi): n = 7, and 3fH_1 (dUtx RNAi): n = 12). p-values: 2fH_1 = 0.0122, 3fH_1 = 0.0005. B Representative image of a nucleus 60 min after 5 Gy irradiation immunostained for anti-yH2Av (yellow), anti-Rad51 (cyan) and anti-mCherry to visualize ph-p-mCherry (magenta). Zoom-in (below) shows a yH2Av focus colocalizing with Rad51, outside a php-mCherry domain. Scale bar = 1 μm. Dotted line outlines nucleus. C Quantification of the number of Rad51 foci per cell in the absence (control, yellow dsRNA, black line) and presence of dUtx dsRNA (orange line). Average number of Rad51 foci per cell is shown at indicated time points. Shade represents SEM of n = 4 (control) or n = 3 (dUtx dsRNA) biological replicates (with ≥10 cells per condition). p-value: 60 min = 0.0067. D Quantification of the localization of Rad51 60 minutes after 5 Gy IR either ‘inside’ a polycomb body or ‘outside’ a polycomb body. Dots represent n = 4 (control) or n = 3 (dUtx dsRNA) biological per condition (with ≥10 cells per condition). Straight line in the violin plot indicates the median and dotted lines indicate quartiles. p-value: outside = 0.0110. (ns) not significant, (*) p-value ≤ 0.05, (**) p-value ≤ 0.01, (***) p-value ≤ 0.001, two-sided unpaired t-test (A, C, D). Source data are provided as Source Data file.