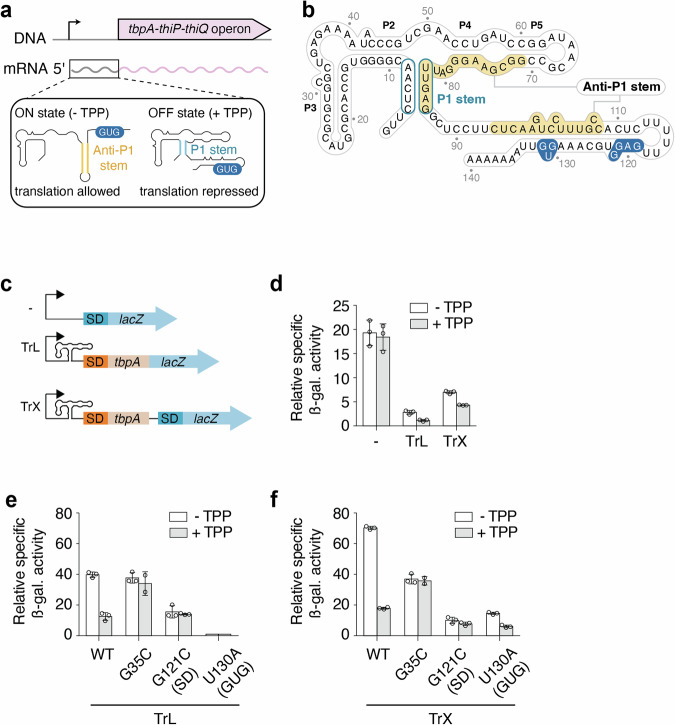
Fig. 1. The regulation mechanism of the E. coli tbpA riboswitch.
a The tbpA riboswitch controls the expression of the tbpA-thiP-thiQ operon that is involved in the transport of thiamin and related metabolites. The riboswitch is located in the 5’ untranslated region and adopts the ON state in the absence of TPP in which the anti-P1 stem is formed, thereby allowing the initiation of translation. However, in the presence of TPP, the OFF state is adopted in which the P1 stem of the aptamer domain is stabilized, therefore repressing translation initiation. The GUG start codon is shown in blue. b Secondary structure of the tbpA riboswitch in the TPP-bound OFF structure. The anti-P1 stem is highlighted in yellow and the P1 stem is circled in blue. The Shine-Dalgarno and GUG start codon are shown in blue. c Schematics depicting the constructs used for ß-galactosidase assays. All fusions contain the natural promoter. The “- fusion” represents the construct in which the riboswitch domain is not present. The translational fusion (TrL) contains 11 codons of tbpA and is directly fused to the lacZ reporter. The transcriptional fusion (TrX) contains an additional Shine-Dalgarno (SD) sequence that allows the translation of the lacZ reporter without being affected by the structure of the riboswitch. d ß-galactosidase assays for the promoter-only (-), translational (TrL) and transcriptional (TrX) fusions. The average values of three independent experiments with standard deviations are shown. ß-galactosidase assays for constructs expressed from an arabinose-inducible promoter (pBAD). Gene expression was assessed for the wild-type (WT), G35C, G121C and U130A constructs in the context of both the translational (TrL) (e) and transcriptional (TrX) (f) fusions. The average values of three independent experiments with standard deviations are shown.