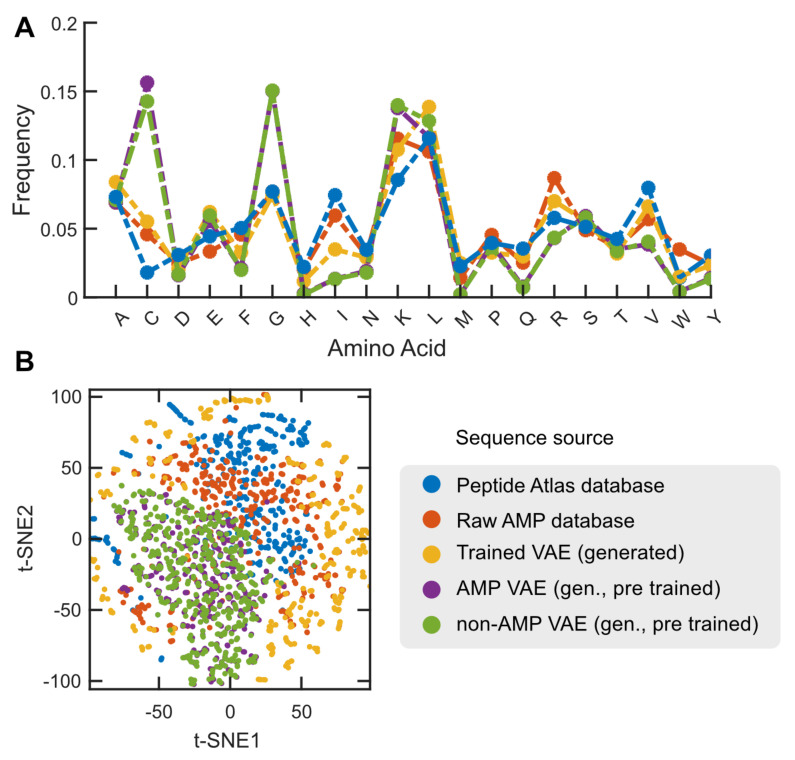
Figure 4.
Visualization of generated antimicrobial peptides by applying VAE approaches. (A). Averagefrequency of amino acids in the studied sequences depends on their source of origin. Sequences created using pre-trained VAEs tend to have slightly more cysteine and glycine instances, regardless of whether the original input was an AMP or not. On the other hand, raw AMPs, potential AMPs identified in the Peptide Atlas, and AMPs generated using VAE trained with AMPs all show similar patterns, except for isoleucine and leucine. In these cases, the peptides generated using VAEs have a lower or higher frequency, respectively (see Table S4 in the Supplementary Materials for more details). (B). Embedding visualization via t-SNE for the numerical representations generated by the ProTrans t5 Uniref pre-trained model for the different sources analyzed. The sequences generated by the VAE trained with AMP sequences show greater dispersion and visual separation compared to other sources, indicating possible new behaviors. This is reflected in the variations in the amino acid properties and frequency. The representations for the potential AMPs generated via the pre-trained VAE exhibit similar behavior. The same is true for the raw AMP sequences and the potential AMPs identified in the Peptide Atlas, consistent with the analysis of the properties and amino acid frequencies.