Figure 1.
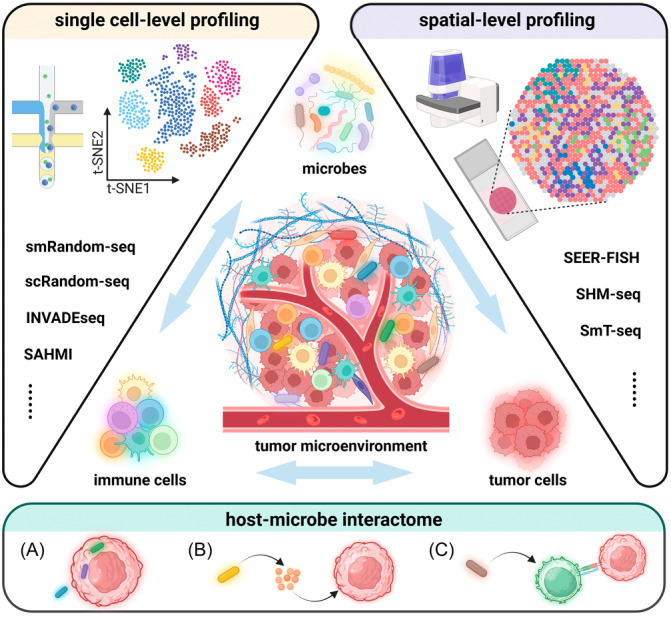
Single‐cell and spatial profiling approaches to decipher the multifaceted host‐microbe interactome. The advent of single‐cell and spatial profiling approaches allows for the precise analysis of individual host and microbial cells. In the context of single cell and spatial‐level profiling, we are able to decipher multi‐modal host‐microbe interactomes that has been overlooked in metagenomic sequencing. For example, (A) microbes may attach to and invade tumor cells to affect their immune phenotypes; (B) microbes can produce multiple substances to modulate TME distantly, including metabolites, extracellular vesicles, and so on; (C) microbes may act as the third party to break the balance between immune cells and tumor cells by regulating the differentiation and activation of immune cells. (Abbreviations: INVADEseq, invasion‐adhesion‐directed expression sequencing; SAHMI, single‐cell analysis of host‐microbiome interactions; SHM‐seq, spatial host‐microbiome sequencing; SmT‐seq, spatial metatranscriptomics sequencing).
