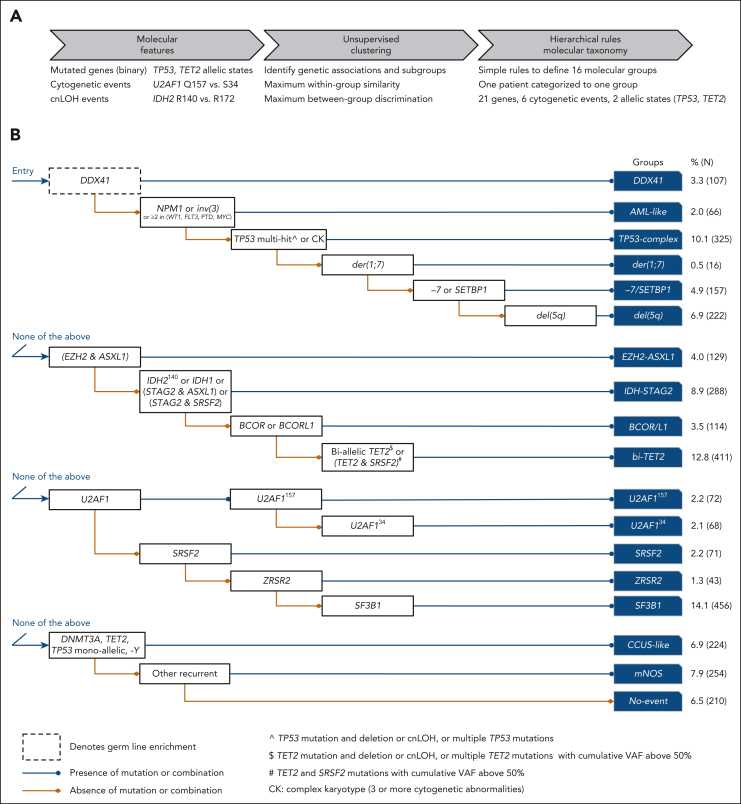
Figure 2.
Derivation of 16 MDS molecular groups. (A) Schematic of the analytical workflow to identify molecular groups. Features were based on the presence or absence of genetic alterations (gene mutations, cytogenetic, and cnLOH events). Distinct allelic states or hot spots were also included based on prior knowledge (TP53 allelic state, IDH2 hot spots) or univariate comutation analysis (TET2 allelic state, U2AF1 hot spots). (B) Rules of co-occurrence and mutual exclusivity of genetic alterations organized in a hierarchical classification tree.