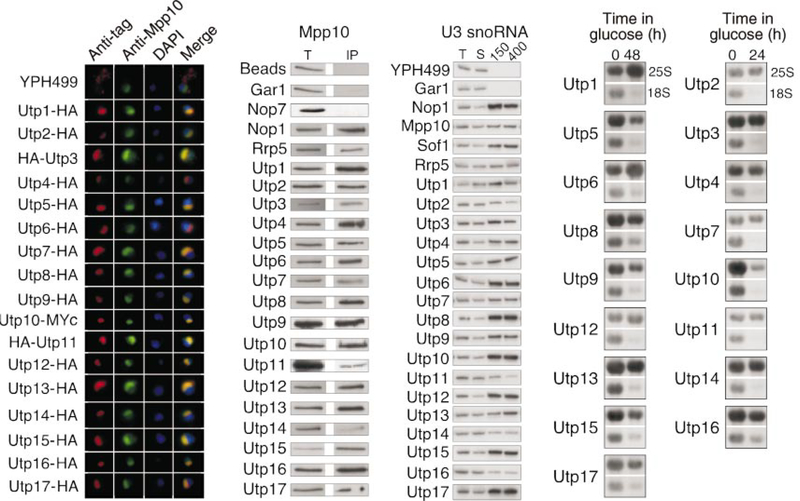
Figure 2.
Function of the new components of the SSU processome. a, The Utp proteins are nucleolar. Anti-HA or anti-Myc antibodies were used to detect the tagged Utp proteins (red), whereas anti-Mpp10 antibodies were used to decorate the nucleolus (green). DAPI was used to stain the nucleus (blue). Utp proteins were triply tagged by HA or Myc. b, The Utp proteins and Rrp5 are complexed with Mpp10. Anti-HA immunoprecipitations performed on cell extracts were analysed for the presence of Mpp10 by western blotting with anti-Mpp10 antibodies. T, total (5% of the input for immunoprecipitation); IP, immunoprecipitate. c, The Utp proteins and Rrp5 are associated with U3 snoRNA. Anti-HA immunoprecipitations were performed on cell extracts and were washed at 150 mM (150) or 400 mM (400) NaCl. RNA was extracted and analysed for the presence of the U3 snoRNA by northern blotting. T, total (20% of the input); S, supernatant (20%). d, The Utp proteins are required for 18S rRNA biogenesis. Yeast strains conditionally expressing Utp proteins were grown in medium containing galactose/raffinose (0 h), but switched to glucose for protein depletion (24 or 48 h). RNA was extracted at the indicated time points and analysed for the presence of 25S and 18S rRNAs by northern blot.