FIGURE 9.
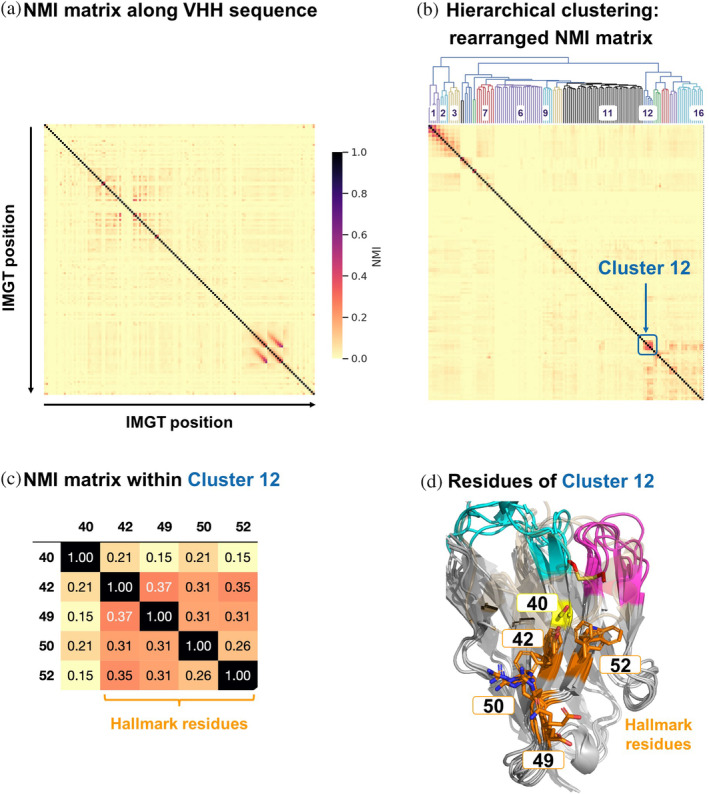
(a) NMI matrix based on normalized Shannon entropies along the VHH sequence (in IMGT numbering). (b) Rearranged NMI matrix, including the dendrogram associated with the hierarchical clustering and the assigned cluster IDs. Cluster 12 is indicated in the matrix. (c) NMI matrix for residues within Cluster 12 that indicate the dependencies between the pairs of amino acid positions. Residues of the classical Hallmark signature are indicated. (d) 3D‐alignment of six PDB structures (PDB codes 1BZQ, 1F2X, 1IEH, 1KXV, 1QD0, 1SJX) with different Hallmark signatures. Key residues are indicated in the same color coding as in Figure 1 and position 40 is indicated in yellow.
