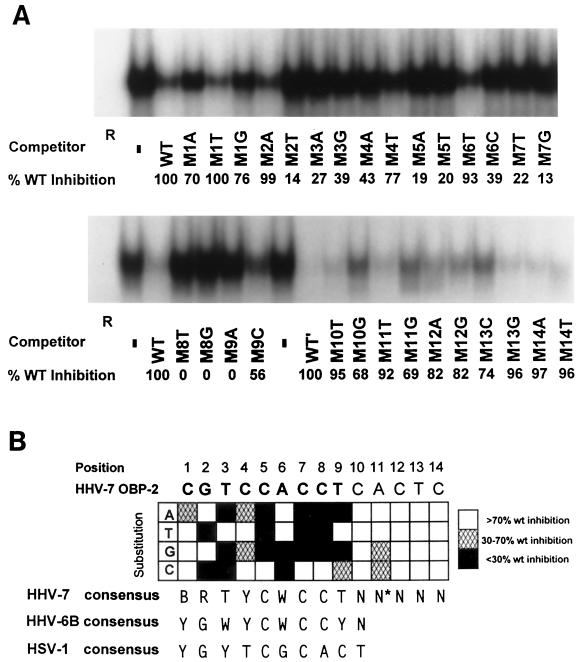
FIG. 8.
Effect of substitutions in the 14-bp core OBP-2 sequence and the resulting OBPH7 consensus recognition sequence. (A) Saturation mutagenesis of the minimal OBPH7 recognition sequence (Table 1, oligonucleotide L0R5). At each position of the 14-bp minimal recognition sequence, oligonucleotides were generated that contained changes to the other three possible base pairs. The label beneath each lane of the gel indicates the position and change made in the competitor used. Sixty-fold molar excesses of unlabeled mutated oligonucleotides were used in competitive EMSA against 32P-labeled 7–2B. The amount of residual shifted radioactivity after competition was quantified by PhosphorImager analysis (Molecular Dynamics). The inhibition of binding of 32P-labeled 7–2B DNA to truncated OBPH7 by each competitor DNA duplex is shown beneath the gel as the percent inhibition relative to the wild-type DNA duplex (WT). The other competitor DNA duplexes shown in panel B were analyzed similarly (data not shown). Positions 10 to 14 were analyzed in the context of the sequence of the longer 7–2Eext oligonucleotide, AATTAGCGTCCACCTCACTCGTAATAGT (WT′), to avoid effects that might be more dependent on proximity to the end of the oligonucleotide than sequence alone. −, no competitor was added. (B) Summary of saturation mutagenesis and the resulting consensus recognition sequence. Open blocks indicate that the given alteration at that position did not result in loss of recognition (competition was at least 70% of WT) and is therefore a permissive change. Hatched and solid blocks indicate that the alteration reduced the binding ability partially or severely, respectively. These designations were based on quantitative trends observed in at least three independent experiments. The asterisk on the N at position 11 indicates that although two of the changes were slightly below the cutoff for recognition, the slight gradient in recognition of all alterations at this position did not enable identification of a clearly preferred sequence. In the consensus sequence, B is a T, G, or C; R is a G or A; Y is a T or C; W is a T or A; and N is any nucleotide.