FIGURE 2.
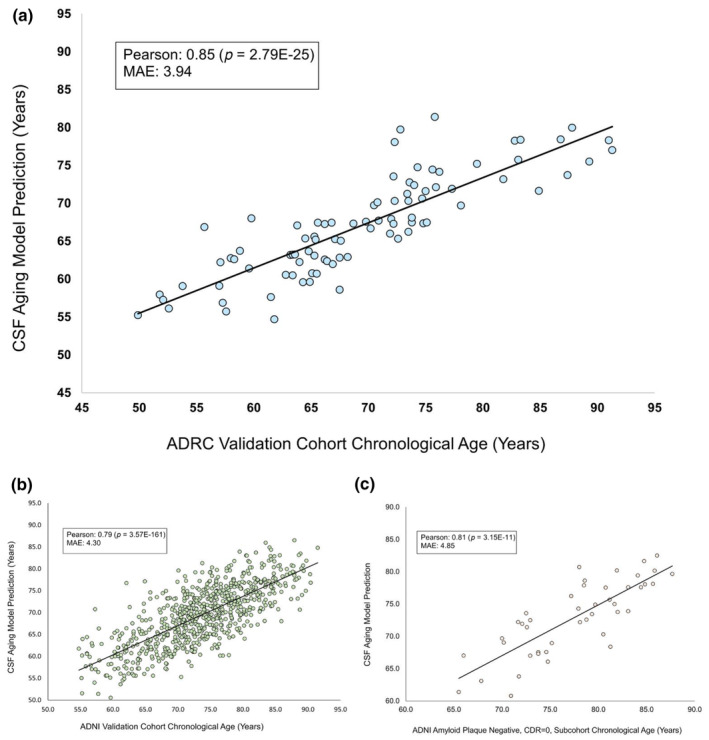
Age predicted via CSF proteomics elastic net model versus chronological age. An elastic net was trained to predict chronological age from SomaLogic CSF proteomics profiles, each containing measurements of 7008 different proteins. (a) The model was trained on 349 cognitively normal, amyloid beta negative, participants from the Knight ADRC, ages 43–91 with a median age of 69. The trained model is shown predicting the ages of an additional 88 cognitively normal participants not included in the training data. The model made use of 1157 protein features to generate predicted CSF age and shows a 0.85 Pearson correlation with chronological age (p = 2.79E‐25) and a mean average error of 3.94 years. (b) The trained model was then validated using CSF proteomics from an additional cohort, the Alzheimer's Disease Neuroimaging Initiative (ADNI) consisting of a new set of 735 individuals. The model validated on this cohort with a Pearson correlation of 0.79 with chronological age (p = 3.57E‐161) and MAE of 4.30 years. (c) As the ADNI cohort included a range of individuals with varying clinical dementia ratings (CDR) and amyloid beta statuses a subcohort was created of 44 ADNI participants who were verified to be cognitively normal and amyloid beta negative. The model was then validated on this subcohort and had a 0.81 Pearson correlation with chronological age (p = 3.15E‐11) and MAE of 4.85 years.
