FIGURE 4.
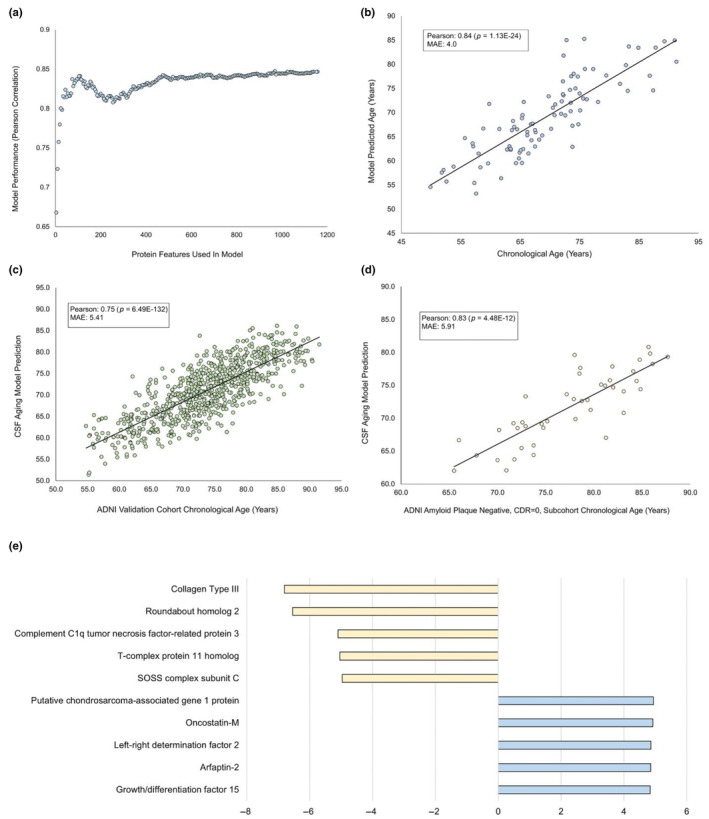
Iterative down‐sampling of proteins to create a minimal CSF aging model. An elastic net was trained on progressively fewer proteins to create a minimal model for predicting chronological age from CSF. Training began with the 1157 protein features identified in the full model. After each round of retraining, proteins were reordered by the absolute values of their new weights, and the bottom 10 proteins were eliminated. (a) The number of proteins used plotted against the Pearson correlation of the model's prediction of ages versus known chronological ages. (b) The results of a 109‐protein “minimal” model predicting age versus known chronological age on the validation cohort. This minimal model had a 0.84 Pearson correlation with chronological age of the ADRC validation group (p = 1.13E‐24) and an MAE of 4.0 (c) The ADRC trained 109‐protein model used to predict chronological ages of the full ADNI validation cohort. The 109‐protein model validated on this cohort with a Pearson correlation of 0.75 with chronological age (p = 6.49E‐132) and MAE of 5.41 years. (d) The ADRC trained 109‐protein model used to predict chronological ages of participants from the ADNI cohort who were verified to be amyloid beta negative and cognitively normal. The 109‐protein model validated on this cohort with a Pearson correlation of 0.83 with chronological age (p = 4.48E‐12) and MAE of 5.91 years. (e) Top 10 protein features weighted by absolute value in the 109‐protein minimal model.
