Figure 4. B11 and B16 protein boosters increase BG18gH B cell SHM and affinity maturation.
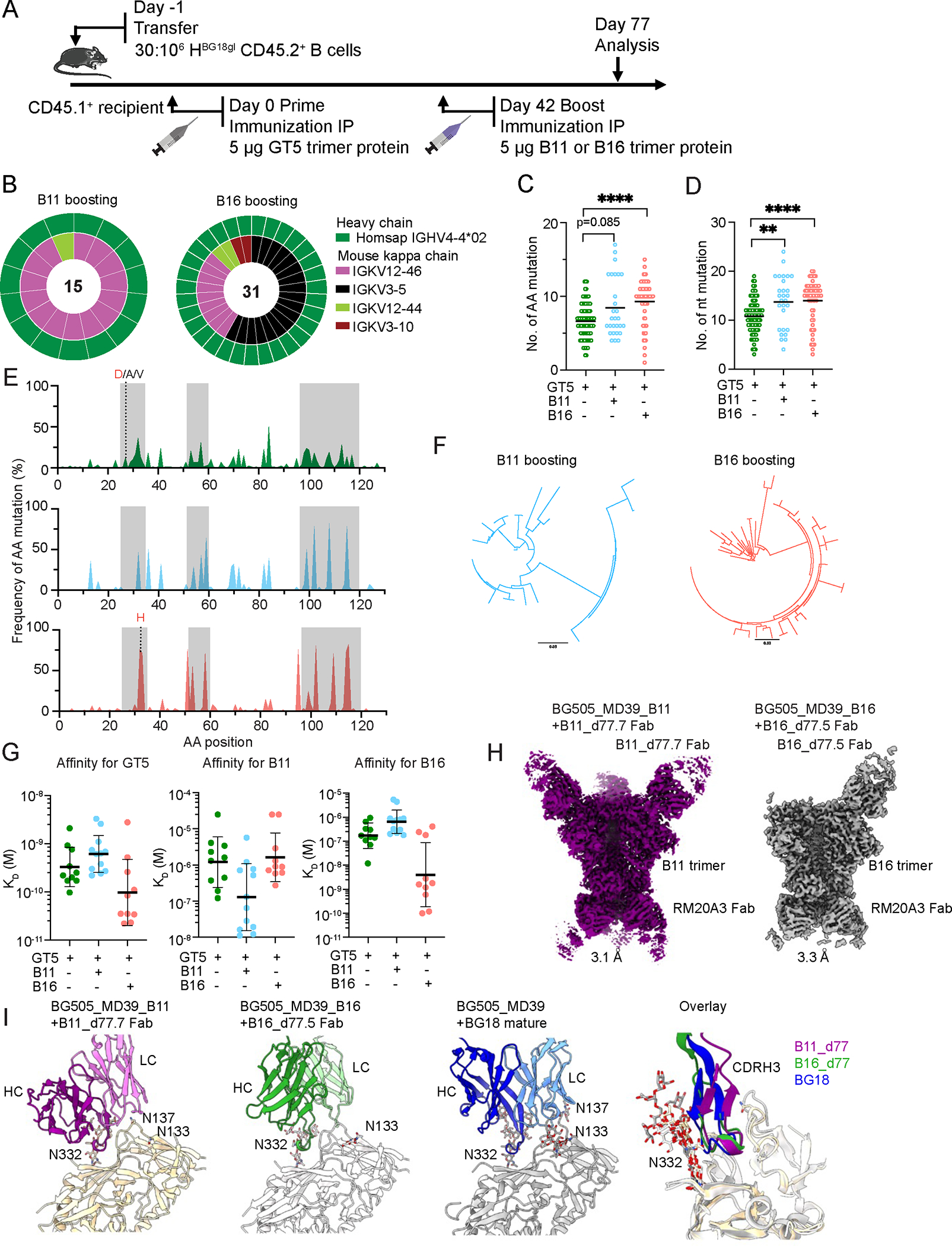
(A) Schematic showing BG18gH B cell adoptive transfer recipients primed with GT5 trimer protein on day 0 and boosted with B11 or B16 trimer protein on day 42. Prime-only samples (green) were collected for analysis on day 42; boost samples (B16 = red; B11 = blue) were collected on day 77. Day 42 prime-only sequences are those presented in Figs. 2G & H and fig. S2G.(B) Nested pie chart showing paired BCR sequences on day 77 from Ag+ (antigen positive) KO− CD45.2+ post-boost B cells. Outer layer represents BG18 IGHV; inner layer represents murine IGKV. (C–D) Heavy chain AA (C) and nt (D) mutations across all sites (source: BCRs sequenced from epitope-specific CD45.2+ B cells on day 42 for prime-only and 77 for boosted mice). Statistical analysis performed using Kruskal-Wallis test. Bars indicate mean. **p <0.01, ****p < 0.0001. (E) Per site heavy chain AA mutation frequency. Red letters represent key BG18 mature or mature-like mutation; grey letters show other mutations on the key residue sites. CDRs are boxed in grey. GT5 prime day 42 (top); B11 boost day 77 (middle); B16 boost day 77 (bottom). (F) Phylogenetic trees of post-boost HCs. Scale (values: 0.03) shows substitutions per site. (G) SPR dissociation constants (KD) for GT5, B11 and B16 trimer binding to Fabs derived from epitope-specific CD45.2+ B cells. Each symbol corresponds to a different Fab and represents one or two measurements. Bars indicate geometric mean and geometric SEM; n=10 or 12. Isolation on day 42 for prime-only and 77 for boosted mice. GT5 prime-only affinity data is as presented in Figs. 2J & 3C. (H) Cryo-EM reconstructions of Fabs derived from epitope specific CD45.2+ BG18 cells at day 77 after either B11 or B16 boost in complex with their respective immunogens. Estimated Fourier Shell Correlation resolutions are listed. (I) Comparison of cryo-EM models of B11 or B16 boost-derived Fabs with mature BG18 (PDB 6dfg). PNGS proximal to the binding epitopes and present in the trimer (B11, B16 or MD39) are highlighted.
