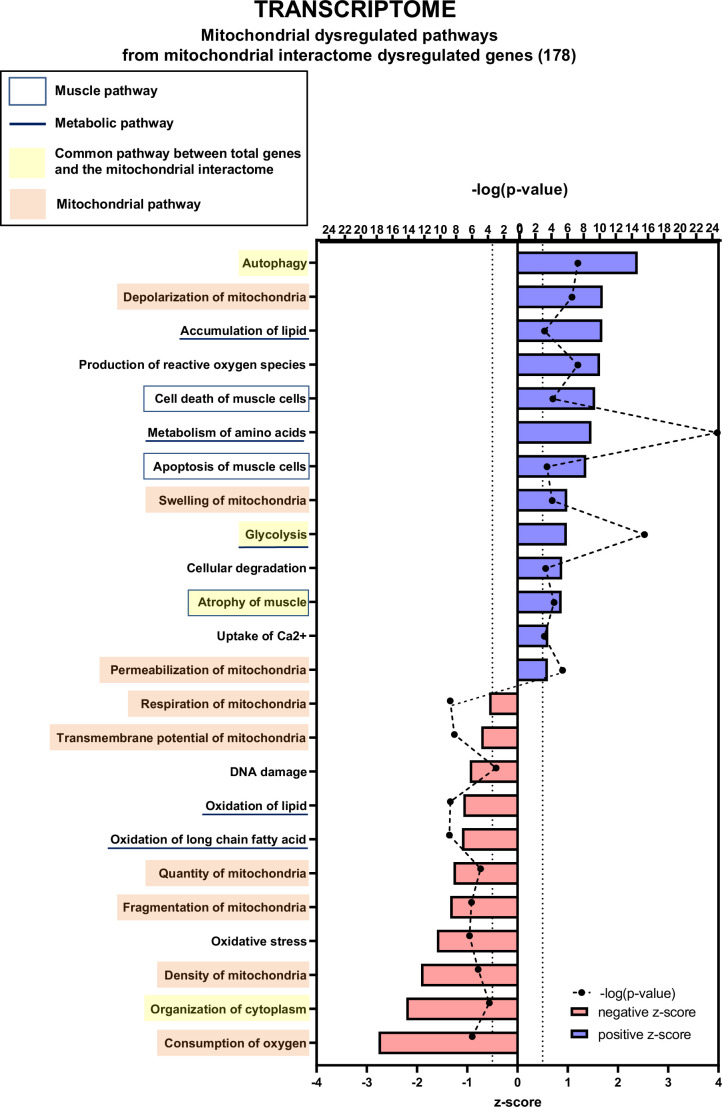
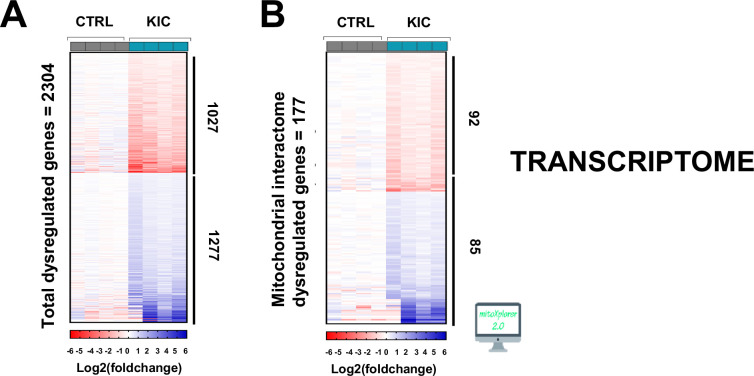
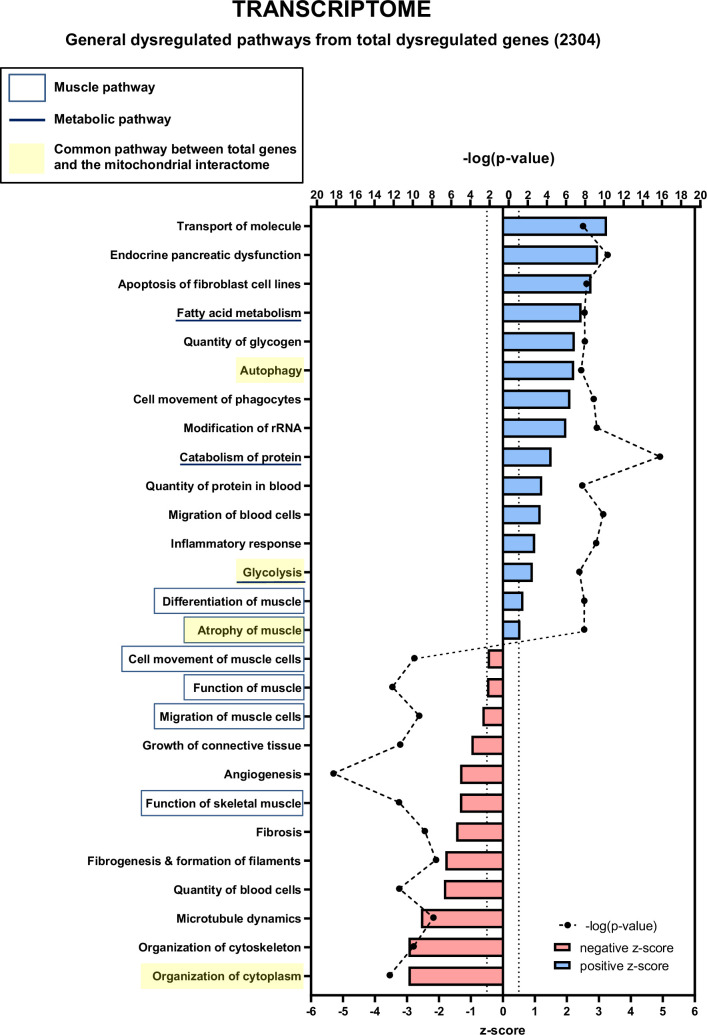
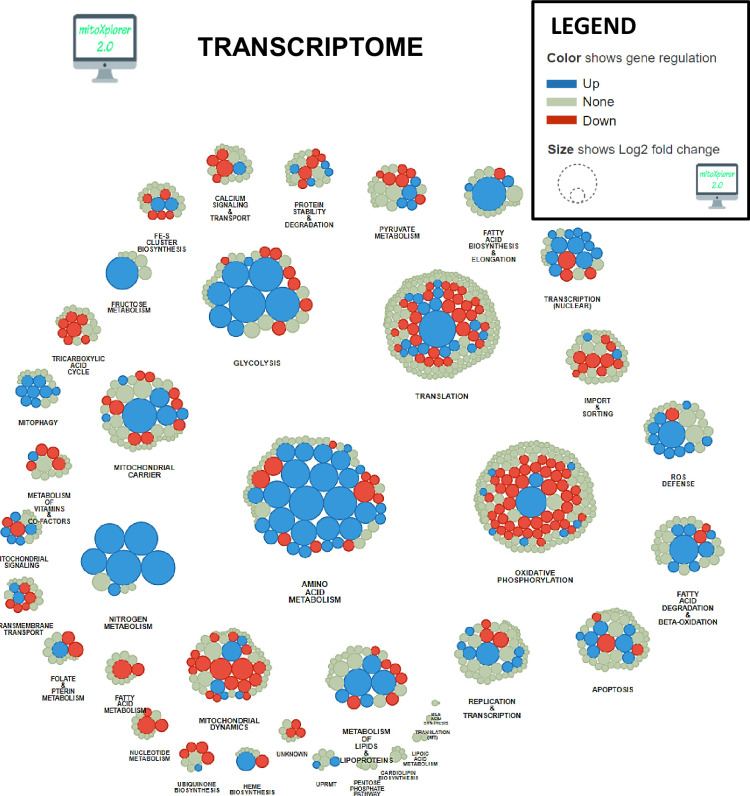
Figure 6. Transcriptomic analysis demonstrates multiple mitochondria-associated gene dysregulations in cancer sarcopenic muscles.
Ingenuity Pathways Analysis (IPA) showing functions, pathways, and diseases significantly dysregulated in gastrocnemius muscle of cancer KIC mice compared to healthy control mice, associated with genes of the mitochondrial interactome extracted from mitoXplorer tool with significant dysregulations (false discovery rate [FDR]>0.05)=178 genes. Each process is represented using z-score>0.5 as upregulated (blue) or downregulated (red), and the significance represented as –log(p-value) with black dots connected with a dotted line. Muscle pathways are framed by a square. Common pathways between total genes and the mitochondrial interactome are highlighted in yellow. Mitochondrial pathways are highlighted in orange. Other metabolic pathways are underlined.