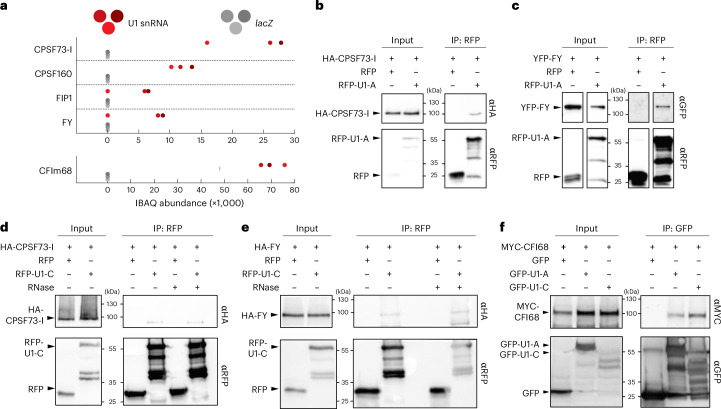
Fig. 2. The U1 snRNP core components, U1-A and U1-C, associate with mRNA cleavage and polyadenylation factors.
a, Abundance of CPAFs in U1-IP–MS experiments. The three red and grey dots represent iBAQ values of three biological replicates using the U1 or the lacZ antisense oligonucleotide, respectively. b,c, U1-A translationally fused to RFP was co-expressed with HA-tagged CFSF73-I (b) or YFP-tagged FY (c) in N. benthamiana plants for transient protein expression. RFP alone served as a negative control. Proteins were isolated and immunoprecipitated using an RFP-affinity matrix. Input and immunoprecipitated fractions (IP) were subjected to protein blot analysis using RFP-, HA- and G/YFP-specific antibodies. Each experiment was repeated two times independently with similar results. d,e, U1-C translationally fused to RFP was co-expressed with HA-tagged CFSF73-I (d) or FY (e) in N. benthamiana plants for transient protein expression. RFP alone served as a negative control. Proteins were isolated and immunoprecipitated using an RFP-affinity matrix in the presence or absence of RNase A. Input and immunoprecipitated fractions (IP) were subjected to protein blot analysis using RFP- and HA-specific antibodies. Each experiment was repeated three times independently with similar results. f, MYC-CFI68 was transiently co-expressed with GFP-U1-A, GFP-U1-C or GFP in N. benthamiana plants. After immunoprecipitation using a GFP-affinity matrix, the isolated proteins were subjected to protein blot analysis. GFP- and MYC-specific antibodies were used for the detection of the tagged proteins. Each experiment was repeated three times independently with similar results.