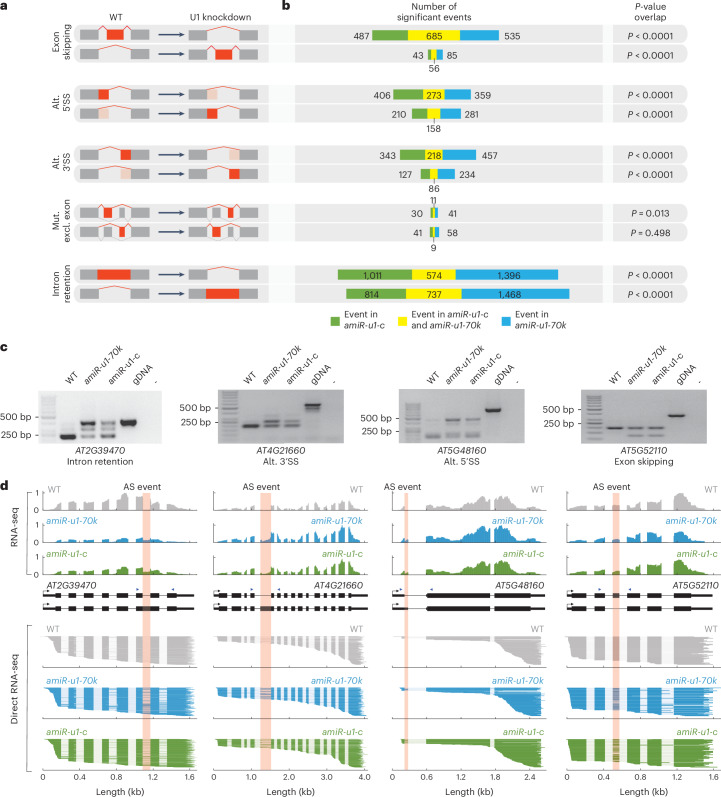
Fig. 4. Knockdown of U1-70K or U1-C causes overlapping splicing defects.
a,b, Changes in the splicing pattern were calculated on the basis of RNA-seq data from WT (a) and amiR-u1-70k and amiR-u1-c (b) plants using rMATS. Splicing changes were subcategorized into exon skipping, alternative (alt.) 5′SS, alternative 3′SS, mutually exclusive exons (mut. excl. exon) and intron retention. A schematic representation of the different splicing changes is shown in a. The numbers of significantly differential alternative splicing events are shown. Significance of overlaps in splicing changes in amiR-u1-70k and amiR-u1-c plants was tested using one-sided hypergeometric overlap test. c, RT–PCR analysis of selected alternative splicing events detected in the RNA-seq dataset. Primers used for amplification were designed to flank the splicing event. Position of primers is depicted by blue arrowheads in d. d, ONT direct RNA-seq reads aligned to the genes that produced alternative spliced RNAs (c). The coverage plot of one representative replicate of the RNA-seq dataset used for rMATS analysis (a,b) is shown. Pink boxes indicate the alternative splicing (AS) events detected by rMATS.