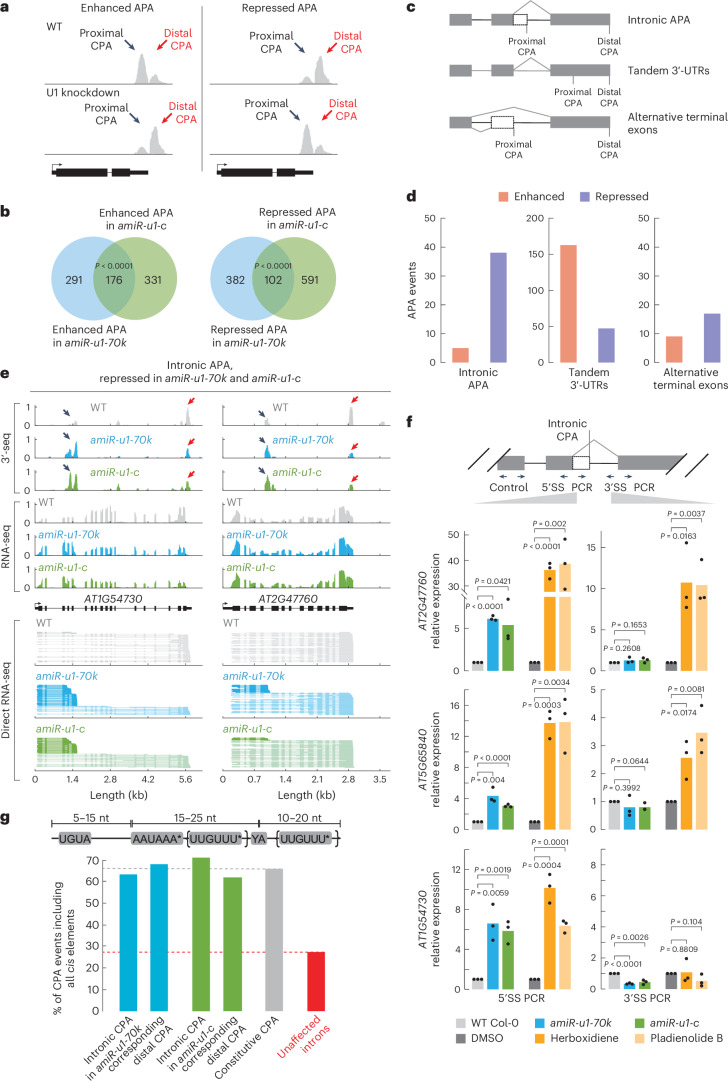
Fig. 5. The U1 snRNP regulates alternative polyadenylation in Arabidopsis.
a, A schematic representation of enhanced and repressed APA events. In enhanced APA events, the proximal CPA site is preferentially utilized. In repressed APA events, the distal CPA site is preferentially utilized. Black arrows indicate the proximal CPA and red arrows indicate the distal CPA. b, Polyadenylation sites were detected by 3′-end sequencing of RNAs (3′-seq) experiments using RNA isolated from 7-day-old WT, amiR-u1-70k and amiR-u1-c seedlings. Venn diagrams depict the overlap of enhanced or repressed APA events in amiR-u1-70k and amiR-u1-c when compared with WT. Significance was tested using a one-sided hypergeometric overlap test. c, A schematic representation of three different types of APA: intronic APA, tandem 3′-UTRs and alternative terminal exons. d, Number of different APA events detected in both amiR-u1-70k and amiR-u1-c plants, when compared with WT. Intronic APA, tandem 3′-UTR and alternative terminal exons were further divided into enhanced and repressed events. e, Two examples of intronic APA events that are repressed in amiR-u1-70k and amiR-u1-c plants. The figure depicts the gene models and the corresponding coverage plots for 3′-seq, RNA-seq and direct RNA-seq. f, RT–qPCR analysis of 7-day-old WT, amiR-u1-70k and amiR-u1-c seedlings, or WT seedling grown in liquid culture for 7 days and treated with DMSO (mock), berboxidiene or pladienolide B. PCR was performed with oligonucleotides spanning the 5′SS or 3′SS and results were normalized to an internal control. The bars indicate the average relative expression in three biological replicates and the dots represent the three independent measurements. A one-sided t-test was applied. g, Analysis of the co-occurrence of three specific cis-elements in different genomic features. Existence of UGUA, AAUAAA (allowing one mismatch except AAAAAA) and a UUGUUU motif (allowing one mismatch except UUUUUU) before or after the cleavage sites were analysed. Genomic features were chosen as follows: introns that are pCPAed in amiR-u1-70k or amiR-u1-c (and the corresponding distal CPA site), CPA sites in genes exhibiting only a single CPA site (constitutive CPA) and all introns that are not pCPAed (unaffected introns).