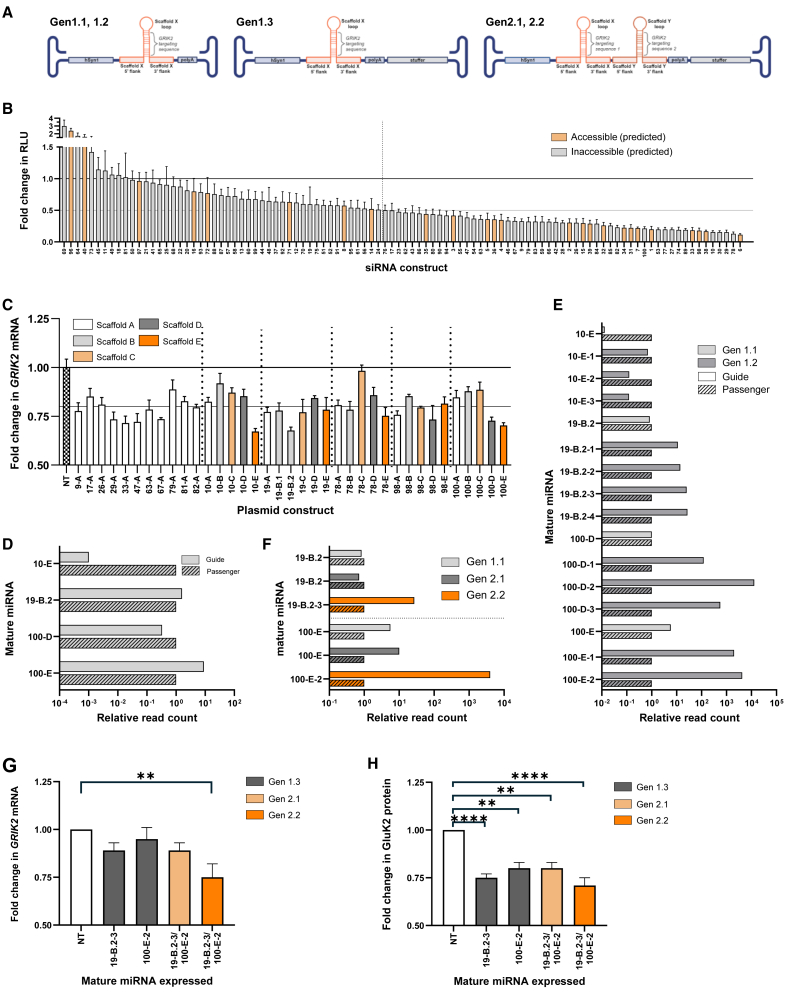
Figure 1.
Design, selection, and optimization of synthetic miRNA constructs capable of efficient GluK2 knockdown with favorable miRNA processing profiles
(A) Schematics depicting the vector genome formats of Gen1.1-.3 and Gen2.1-.2 constructs. (B) Quantification of firefly luciferase(ffluc)-GluK2 reporter knockdown mediated by siRNA and dual reporter plasmid transfection in 293 cells. Data are reported as ffluc RLU from co-transfection of experimental siRNAs with ffluc-GluK2 reporter relative to ffluc RLU from co-transfection of negative control siRNA with ffluc-GluK2 reporter, after normalization to control renilla luciferase RLU (reported as mean ± SEM). (C) Quantification of GRIK2 mRNA levels following miRNA plasmid construct transfection of Gen1.1 constructs into iCell GlutaNeurons, relative to lipid only transfection control (NT), as measured by real-time qPCR and reported as mean ± SD. (D) Quantification of relative levels of mature guide and passenger strand expression from top GRIK2-targeting Gen1.1 constructs as measured by smRNA-seq following plasmid transfection of iCell GlutaNeurons. (E) Assessment of the impact of optimization efforts on G:P by smRNA-seq following Gen1.1 and Gen1.2 plasmid transfection of N2A cells. (F) Assessment of the impact of miRNA concatemerization and addition of a stuffer sequence on G:P by smRNA-seq following plasmid transfection of N2A cells. (G) Quantification of GRIK2 mRNA levels by real-time qPCR (normalized to GAPDH) following transduction of C57Bl6/J mouse cortical neurons with AAV9 vectors packaging Gen1.3, Gen2.1, and Gen2.2 constructs; data are reported as mean ± SEM, NT = non-transduced. (H) Quantification of GluK2 protein levels by western blot (normalized to β-actin) following transduction of C57Bl6/J mouse cortical neurons with AAV9 vectors packaging Gen1.3, Gen2.1, and Gen2.2 constructs. ∗∗p < 0.01, ∗∗∗∗p < 0.0001 one-way ANOVA, followed by Dunnett’s multiple comparison. Data are reported as mean ± SEM, NT = non-transduced.