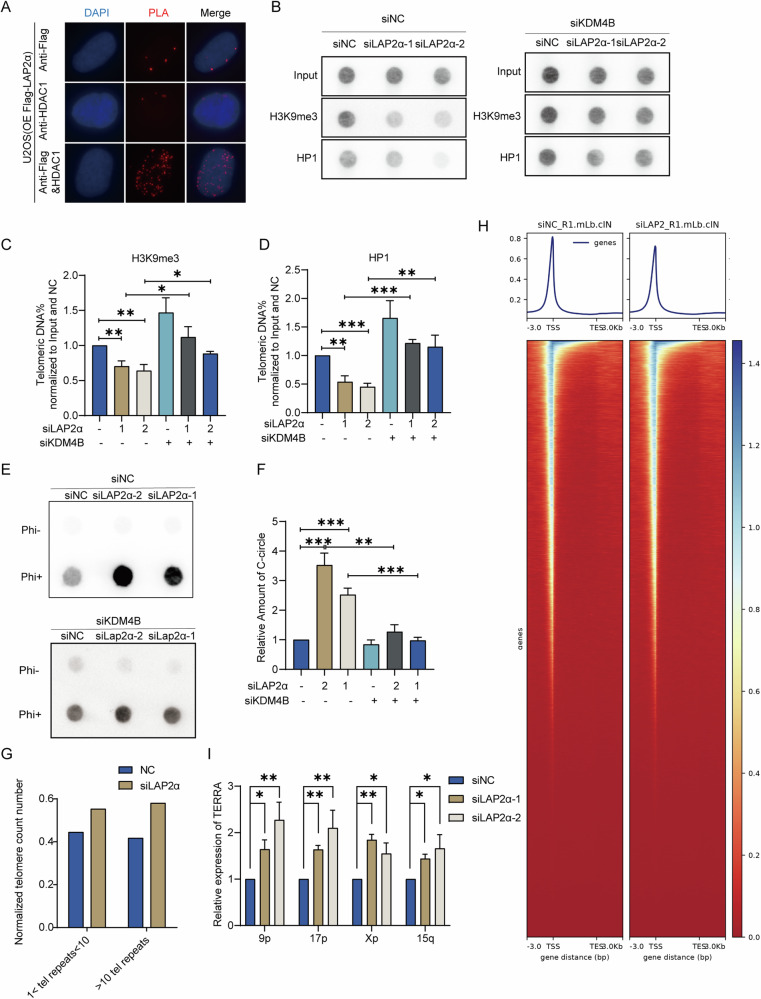
Fig. 5. Lack of LAP2α decreased the heterochromatin status of telomere.
A PLA assay was used to analyze the interaction between LAP2α and HDAC1 in U2OS over-expressing Flag-LAP2α. Red dots represent PLA signals. B U2OS cells were transiently transfected with control siRNA or LAP2α siRNA for 48 h and then transfected with control siRNA or KDM4B siRNA for 48 h, cells were harvested and subjected to ChIP experiments using antibodies raised against trimethylated H3K9, HP1 and control IgG binding to telomeres. Telomere repeat DNA was visualized by dot blot and probed with a telomere-specific probe. C, D Quantification of H3K9me3 and HP1 binding to telomeres at least three different ChIP assays represented by panel (B). Error bars represent the mean ± SEM of three independent experiments. Two-tailed unpaired Student’s t-test was used to calculate p-values. *p < 0.05; **p < 0.01; ***p < 0.001. ChIP DNA signals were normalized to input DNA signal and to siNC-treated cells. E C-circle assay in U2OS cells transfected with LAP2α siRNA and/or KDM4B siRNA. F Quantification of the relative amount of C-circle. Error bars represent the mean ± SEM of three independent experiments. Two-tailed unpaired Student’s t-test was used to calculate p-values. **p < 0.01; ***p < 0.001. G Copy number normalized ATAC-seq counts mapped to telomere regions. H Average profiles of ATAC-seq peaks in control siRNA or LAP2α siRNA treated U2OS cells at transcription start site (TSS) ± 3 kb (top). Heatmap of densities of ATAC-Seq peaks of genes in control siRNA or LAP2αsiRNA treated U2OS cells (bottom). I qRT-PCR analysis of TERRA levels in the indicated U2OS cells. TERRA levels were detected using primers specific for TERRA transcribed from subtelomeres of human chromosome 9P, 17P, Xp and 15q, as indicated. Error bars represent the mean ± SEM of three independent experiments. Two-tailed unpaired Student’s t-test was used to calculate p-values. *p < 0.05; **p < 0.01. Data are represented as mean ± SEM of three or more independent experiments, and *p < 0.05; **p < 0.01; ***p < 0.001 (Student’s t-test).