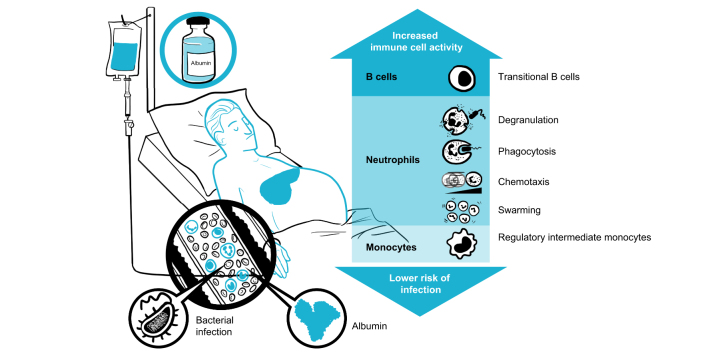
Abstract
Background & Aims
Patients with acutely decompensated (AD) cirrhosis are immunocompromised and particularly susceptible to infections. This study investigated the immunomodulatory actions of albumin by which this protein may lower the incidence of infections.
Methods
Blood immunophenotyping was performed in 11 patients with AD cirrhosis and 10 healthy volunteers (HV). Bulk and single-cell RNA sequencing (scRNA-seq) and flow cytometry were performed in peripheral blood mononuclear cells (PBMCs) from 20 patients with AD cirrhosis and 34 HV exposed to albumin. Albumin’s effects on degranulation, phagocytosis, chemotaxis, and swarming of neutrophils from six patients with AD cirrhosis and nine HV were assessed by measuring myeloperoxidase enzymatic activity, the engulfment of fluorescent-labeled Escherichia coli and zymosan, and interactions of neutrophils with Candida albicans at single-cell resolution in microfluidic chambers, respectively. Whole blood RNA sequencing (RNA-seq) analyses were performed in 49 patients admitted for severe AD cirrhosis, of whom 30 received albumin during hospitalization.
Results
Compared with HV, patients with AD cirrhosis showed severe lymphopenia and defective neutrophil antimicrobial function. Bulk and scRNA-seq analyses revealed significantly (false discovery rate [FDR] <0.05) increased signatures related to B cells, myeloid cells, and CD4+ T cells in PBMCs incubated with albumin. Changes in the B cell population were confirmed by flow cytometry. Neutrophils exposed to albumin also exhibited augmented chemotactic and degranulation responses, enhanced phagocytosis, and increased pathogen-restrictive swarming. RNA-seq data analysis in patients who had received albumin revealed specific upregulation of signatures related to B cells and neutrophils together with transcriptional changes in CD4+ T cells (FDR <0.05).
Conclusions
The finding that albumin promotes the transcriptional reprogramming and expansion of the B cell compartment and improves neutrophil antimicrobial functions indicates mechanisms that may lower the incidence of infections in patients with severe AD cirrhosis receiving albumin therapy.
Impact and implications:
Patients with acutely decompensated cirrhosis receiving albumin as treatment have a lower incidence of infections. The reason for this protection is currently unknown, but the present study provides data that support the ability of albumin to boost the antimicrobial functions of immune cells in these patients. Moreover, these findings encourage the design of controlled clinical studies specifically aimed at investigating the effects of albumin administration on the immune system.
Keywords: Multiorgan failure, Inflammation, Immunosuppression, Gene expression
Graphical abstract
Highlights:
-
•
Patients with acutely decompensated cirrhosis are immunocompromised and have higher predisposition to infections.
-
•
scRNA-seq and FACS analyses demonstrate the expansion of the B-cell compartment in mononuclear leukocytes exposed to albumin.
-
•
Polymorphonuclear leukocytes exposed to albumin also exhibit enhanced neutrophil antimicrobial functions.
-
•
Analysis of RNA-seq data in patients receiving albumin shows upregulated signatures related to B cells and neutrophils.
-
•
These findings provide mechanisms by which albumin reduces the incidence of infections in patients with acutely decompensated cirrhosis.
Introduction
Acute decompensation is the most frequent cause of nonelective hospital admission of patients with cirrhosis, and its most severe form, acute-on-chronic liver failure (ACLF), is characterized by hepatic and/or extrahepatic organ failures and high risk of short-term death.1,2 These patients exhibit intense systemic inflammation, indicated by leukocytosis,2,3 elevated blood levels of cytokines, inflammatory lipid mediators, and C-reactive protein (CRP).1,[3], [4], [5] These patients also exhibit immunosuppression, indicated by decreased responses of specific monocyte subsets to bacterial products,6,7 decreased capacity to kill microbes,6,8,9 and reduced lymphocyte function.3 Immunosuppression predisposes patients with acutely decompensated (AD) cirrhosis to secondary infections and re-escalation of end-organ dysfunction and mortality.10 To date, there are no therapies to prevent or treat patients with AD cirrhosis who are immunocompromised or have an impaired immune response. This unmet need is more striking given the ramping prevalence of patients with AD cirrhosis infected with Gram-positive or Gram-negative bacteria resistant to antibiotics.11
Albumin therapy is common in patients with AD cirrhosis, including patients treated with large-volume paracentesis and patients with spontaneous bacterial peritonitis (SBP) or hepatorenal syndrome–acute kidney injury (HRS-AKI).12 Recently, the ANSWER study has demonstrated a lower incidence of SBP and other infections in patients with AD cirrhosis receiving long-term weekly administration of albumin.13 In addition, albumin administration has emerged as an efficacious anti-inflammatory therapy, reducing cytokine levels in patients with AD cirrhosis and ACLF.14 Although albumin pleiotropic effects have classically been attributed to its oncotic and antioxidant properties,15 recent data on leukocytes from patients with AD cirrhosis have shown that albumin modulates cytokine production in response to bacterial DNA by interacting with the endosomal TLR9 signaling pathway.16 The albumin immune restorative effects have also been linked to inactivation of the immunosuppressive effects of PGE2.8 However, at present, a comprehensive and deep understanding of the immunomodulatory actions of albumin in the setting of AD cirrhosis is still lacking.
To assess this, we designed in vitro experiments and performed bulk and single-cell RNA-sequencing (scRNA-seq) and functional assays in peripheral blood mononuclear cells (PBMCs) and neutrophils from patients with AD cirrhosis and healthy volunteers (HV). Confirmatory in vivo whole blood RNA sequencing (RNA-seq) studies were performed in 30 patients with severe AD cirrhosis receiving albumin therapy and in 19 non-albumin-treated patients with AD cirrhosis from the PREDICT study.17 The results of this investigation revealed that albumin induces specific immune cell gene signatures primarily related to the revitalization of B cell and neutrophil defensive functions in patients with AD cirrhosis. These findings expand our understanding of the mechanisms whereby albumin lowers the rate of infections in patients with cirrhosis and improves survival.
Patients and methods
Patients
For immunophenotyping and in vitro experiments in PBMCs, peripheral blood was obtained from 20 patients with AD cirrhosis recruited at the Liver Intensive Care Unit of the Hospital Clínic of Barcelona and 34 HV from the Barcelona Hospital Clínic Blood Bank (Table S1). For neutrophil experiments, peripheral blood was obtained from six patients with AD cirrhosis and nine age-matched HV (Table S2). Whole blood bulk RNA-seq was performed in 10 HV and 49 patients with AD cirrhosis from the PREDICT study,17 of whom 30 had received albumin therapy (see Supplementary methods for selection criteria). All patients provided written informed consent to participate, and approval was granted by the Ethics Committee of the Hospital Clínic of Barcelona (#HCB/2016/0710, #HCB/2018/0899 and #HCB/2015/0427).
Immunophenotyping and isolation of PBMCs and neutrophils
PBMC incubations
PBMCs (1.5–3.0 × 106 cells/ml) seeded in RPMI 1640 medium were incubated with either human serum albumin (HSA, Albutein®, Grifols, Barcelona, Spain), recombinant albumin expressed in Oryza sativa (Sigma-Aldrich, St Louis, MO, USA) (both at 15 mg/ml), or a vehicle control for 24 h at 37 °C in a 5% CO2 incubator. For comparison, experiments with the iso-oncotic compound mannitol (Sigma-Aldrich) (15 mg/ml), albumin-depleted FBS (15% total volume), and IgG from human serum (Sigma-Aldrich) (15 mg/ml) were also performed. Albumin-depleted FBS was prepared using the Pierce™ Albumin Depletion Kit, and depletion was verified using the bicinchoninic acid assay. In some experiments, PBMCs were incubated with the neonatal Fc receptor (FcRn) blocking antibody ADM31 (Aldevron, Fargo, ND, USA) (10 μg/ml) before the addition of albumin.
Bulk RNA-seq in whole blood
Isolation of total RNA from whole blood collected in Tempus tubes, assessment of RNA concentration and integrity, library preparation, sequencing, and processing of RNA-seq data are described in the Supplementary methods.
Bulk RNA-seq and real-time PCR in PBMCs
scRNA-seq in PBMCs
PBMCs (1.5 × 106 cells/ml) seeded in RPMI 1640 medium were incubated with either HSA (15 mg/ml) or the vehicle (culture medium) for 2 h at 37 °C in a 5% CO2 incubator. At the end of the incubation period, cells were rapidly (within 30 min) transferred on ice to the CNAG Single Cell Genomics platform (see Supplementary methods).
Neutrophil degranulation and phagocytosis assays
Neutrophil chemotaxis and swarming assays
A chemotaxis–phagocytosis assay in microfluidic arenas18 was used to test the role of HSA on the ability of neutrophils to migrate directionally toward and to phagocytose Candida albicans yeast. A swarming assay was used to test the contribution of HSA to the ability of neutrophils to contain the growth of Candida clusters. To this purpose, arrays of Candida-adherent spots (200-μm diameter) were printed using a microarray printing platform (Picospotter PolyPico, Galway, Ireland) and a solution of poly-l-lysine (Sigma-Aldrich) (see Supplementary methods).
Results
Characterization of the blood immune cell landscape in patients with AD cirrhosis
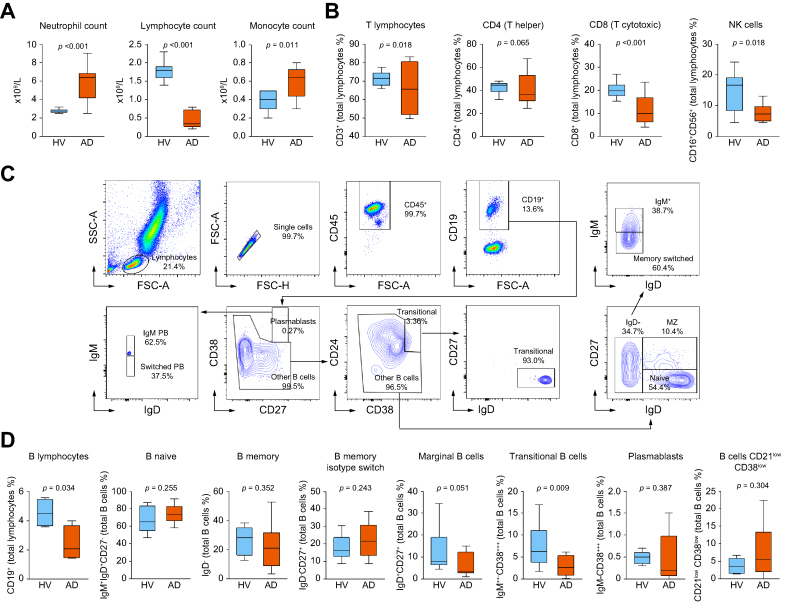
The peripheral immune cell landscape was characterized by flow cytometry in a representative population of patients with AD cirrhosis in whom HSA administration was indicated as standard of care. Compared with HV, patients with AD cirrhosis showed neutrophilia, severe lymphopenia, and monocytosis (Fig. 1A). Lymphopenia affected CD8+ lymphocytes and natural killer (NK) cells (Fig. 1B). The analysis of CD45+CD19+ B lymphocytes, which included the study of distinct B cell subsets defined using specific gating strategies (Fig. 1C), also revealed a decline of B and transitional B cells in patients with AD cirrhosis, suggesting impaired B-cell development and maturation (Fig. 1D).
Fig. 1.
Peripheral immune cell landscape of patients with AD cirrhosis.
(A) Peripheral blood cell counts in 10 HV and 11 patients with AD cirrhosis. (B) Peripheral blood immunophenotyping of T cells, CD4+ and CD8+ T cells, and NK cells in HV and patients with AD cirrhosis. (C) Scatterplots of the flow cytometry analysis showing the gating strategy to identify the different B cell subtypes, including plasmablasts (CD45+CD19+CD38++CD27++), transitional (CD45+CD19+CD38++CD24+IgD+CD27-), naïve (CD45+CD19+IgD+CD27-CD24lowIgM+), marginal (CD45+CD19+IgD+CD27+CD24highIgM++), and switched (CD45+CD19+IgD-CD27+/-IgM-). (D) Proportions of B cells and subpopulations in the peripheral blood of HV and patients with AD cirrhosis. Box and whisker graphs in panels A, B, and D represent the median (IQR). Lower and upper box borders indicate the 25th and 75th percentiles, respectively. Lines within each box indicate median percentage. Whiskers above and below each box indicate maximum and minimum values, respectively. Significance between groups was obtained using Wilcoxon–Mann–Whitney tests. AD, acutely decompensated; HV, healthy volunteers; NK, natural killer.
In vitro response of mononuclear leukocytes from patients with AD cirrhosis to HSA
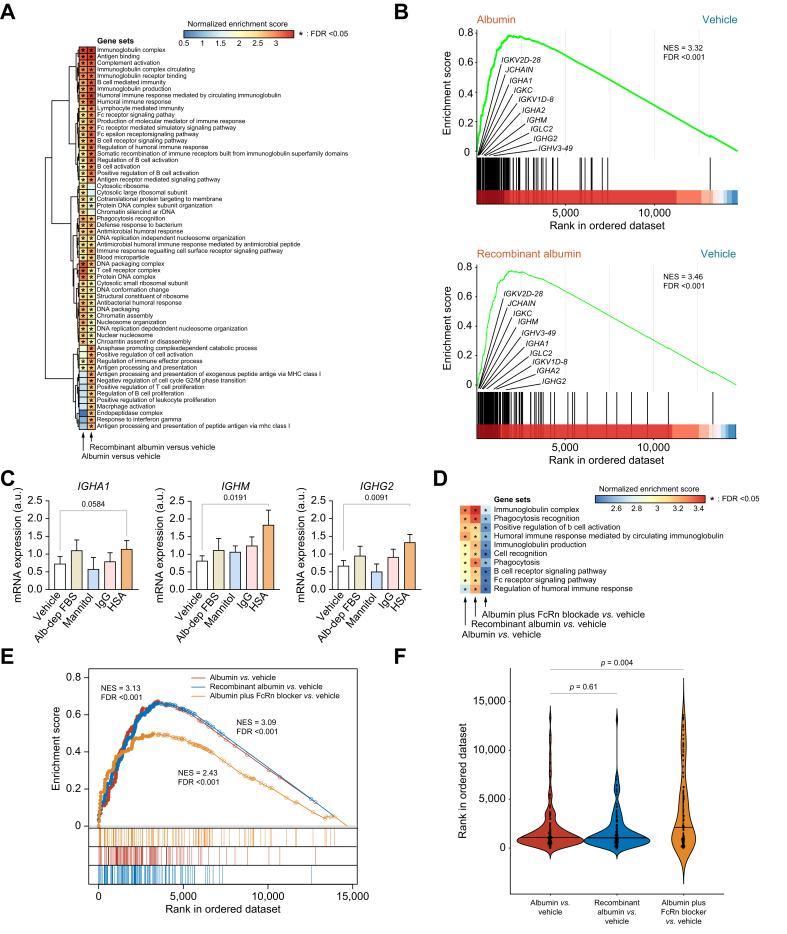
To investigate the direct effect of HSA on immune cells avoiding any confounding factor present in the patient’s bloodstream, we first performed experiments in vitro in PBMCs isolated from patients with AD cirrhosis. To accurately assess the global transcriptional changes elicited by albumin, we performed bulk RNA-seq and identified differentially expressed (DE) gene sets by gene set enrichment analysis (GSEA). This analysis identified two clusters of gene sets related to B-cell-mediated immunity, B-cell activation, immunoglobulin complexes, immunoglobulin receptor binding, immunoglobulin production, immune response mediated by immunoglobulins, and the B cell receptor signaling pathway that were significantly enriched in PBMCs incubated with HSA (Fig. 2A). Among the gene clusters enriched by HSA, we also identified three gene sets related to the Fc receptor signaling pathway (Fig. 2A). HSA also induced the enrichment of two additional gene set clusters, one related to T cells and DNA and protein complexes and the other to antimicrobial responses, phagocytosis recognition, and defense response against bacteria (Fig. 2A). To exclude the possibility that the effect of HSA on PBMCs could be caused by the presence of stimulatory serum factors bound to its molecule and retained during its manufacturing, we repeated the experiments using recombinant human albumin from Oryza sativa. These experiments confirmed that the activation of the B cell transcriptome by albumin is an intrinsic property of this molecule (Fig. 2A). Similar results were observed in PBMCs isolated from HV, indicating that the effect of albumin on B cells was not exclusive to patients with cirrhosis (Fig. S1). Moreover, enrichment analysis identified several leading-edge genes coding for immunoglobulins increased by both HSA and recombinant albumin (Fig. 2B). To assess whether the effect of HSA on immunoglobulin expression was related to its oncotic properties, we compared the effects of HSA with those of the iso-oncotic control mannitol, IgG, and albumin-depleted FBS and demonstrated that leading-edge immunoglobulin genes were only upregulated by HSA (Fig. 2C).
Fig. 2.
Changes in the transcriptional landscape of PBMCs isolated from patients with AD cirrhosis and incubated in vitro with HSA.
(A) GSEA was run on RNA-seq data to generate ranked lists of genes for the two following pairwise comparisons vs. vehicle: HSA and recombinant albumin (both at 15 mg/ml). Color gradient corresponds to increasing values of the NES of gene sets from the less (in blue) to the most (in red) upregulated. Asterisks in the heat map indicate FDR <0.05. (B) GSEA enrichment plots of the immunoglobulin complex gene set in two comparisons vs. vehicle: HSA (top) and recombinant albumin (bottom). The hash plot under GSEA curves shows where the members of the gene set appear in the ranked list of genes. The genes shown on the plots are representative of leading-edge genes (i.e. top-scoring genes). (C) Changes in the expression of three representative leading-edge genes coding for immunoglobulins in response to the vehicle control, 15% albumin-depleted FBS (Alb-dep FBS), mannitol (15 mg/ml), IgG (15 mg/ml), and HSA (15 mg/ml). Significant differences between groups were assessed using t tests. (D) Heat map of the NES for the 10 representative B cell‒related gene sets obtained for each of the following three comparisons vs. vehicle: HSA, recombinant albumin, and FcRn blocker (10 μg/ml) plus HSA vs. vehicle. (E) Enrichment plots of the immunoglobulin complex in the three GSEA comparisons described in D. The leading-edge genes are indicated by solid dots. The hash plots under GSEA curves show where the members of the gene set appear in each of the three ranked lists of genes. (F) Rank order of each gene of the immunoglobulin complex gene set. The higher in the rank, the lower the importance. Values of p are ranked from Kruskal–Wallis tests, followed by Mann–Whitney U tests. All FDR values are computed with adjustments for multiple testing and gene set size in the GSEA analysis. AD, acutely decompensated; FcRn, neonatal Fc receptor; FDR, false discovery rate; GSEA, gene set enrichment analysis; HSA, human serum albumin; HV, healthy volunteers; NES, normalized enrichment score; PBMC, peripheral blood mononuclear cell; RNA-seq, RNA sequencing.
FcRn is implicated in the B cell response to HSA
Having shown that albumin enhances the expression of gene sets related to the Fc receptor signaling pathway, we explored whether the B cell stimulatory effect of HSA was mediated by its binding to FcRn, an Fc receptor that binds albumin with high affinity.19 The normalized enrichment score (NES) for the immunoglobulin complex in PBMCs incubated with HSA in the presence of an FcRn-blocking antibody was lower than that in PBMCs incubated with HSA alone (Fig. 2D and E). Consistent with this, immunoglobulin genes showed a less preferred position in the ordered ranked list of genes in PBMCs incubated with HSA in the presence of the FcRn blocker than in PBMCs incubated with HSA alone (Fig. 2F). Although these findings suggest some implication of the FcRn receptor in the transcriptional reconfiguration of B cells elicited by HSA, other receptors or mechanisms are also likely involved in this transcriptional rearrangement.
Profile of the in vitro effects of HSA at the scRNA-seq level
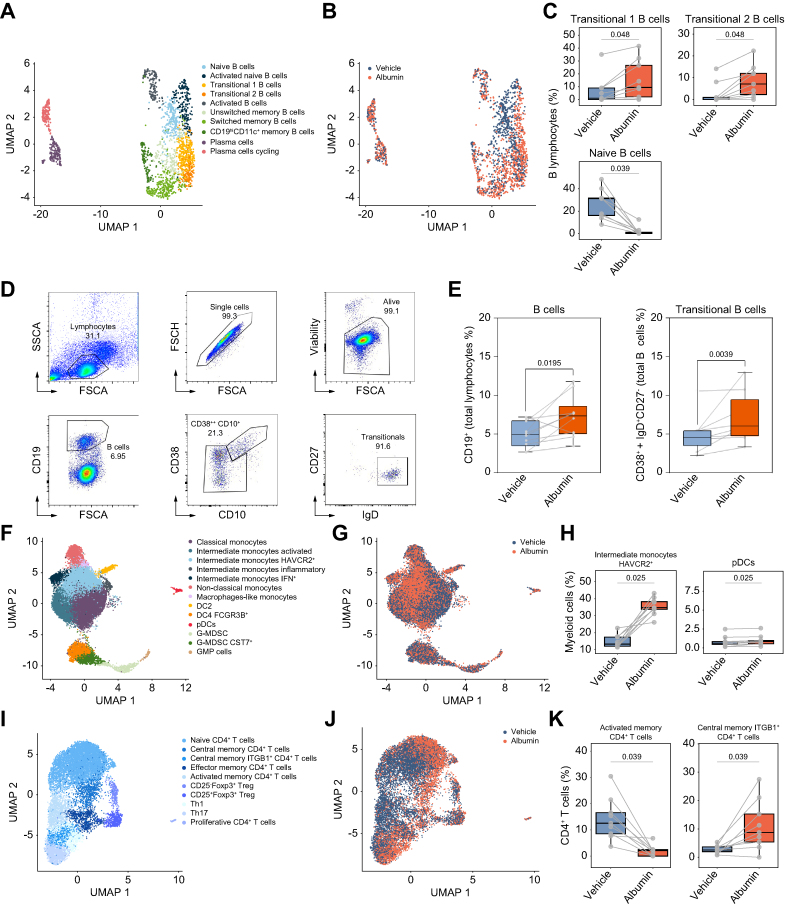
We next used scRNA-seq to gain a deeper insight into the multitiered complexity of the cellular composition of PBMCs exposed in vitro to HSA for 2 h. We jointly analyzed 66,064 human PBMCs from nine patients with AD cirrhosis clustered into B lymphocytes, T lymphocytes, and myeloid cells. Each lineage was subclustered to define fine-grained cell populations using canonical gene markers (Figs. S2 and S3). We then analyzed 1,946 B lymphocytes and identified 10 principal populations, including two subpopulations that acquired a transitional-like B cell profile characterized by a higher expression of CD79B, IGHD, and TCL1A (transitional 1 B cell) along with a higher expression of CD55 (transitional 2 B cell) (Fig. 3A). HSA increased the frequency of B cells expressing a transitional-like profile while reducing naive B cells (Fig. 3B and C). To further confirm that HSA induced transcriptional changes resembling those of transitional-like B cells, we assessed the expression of specific genes described by Steward et al.20 This analysis proved the HSA-induced expression of TCL1A, VPREB3, PCDH9, IGHM, and IGHD genes in transitional B cells (Fig. S4). Importantly, HSA-induced increased frequency of transitional-like B cells observed at the transcriptional level was confirmed by flow cytometry at 24 h (Fig. 3D and E). Interestingly and consistent with the essential role of albumin in in vitro B-cell growth,21 a significant increase in the B cell population was detected by flow cytometry (Fig. 3E). However, albumin did not expand any other B cell subset (Fig. S5A). In addition, no effects on B cells were observed in response to albumin-depleted FBS, mannitol, or IgG (Fig. S5B).
Fig. 3.
scRNA-seq and flow cytometry analyses identify specific immune cell changes in PBMCs exposed in vitro to HSA.
(A) UMAP of 1,946 patients’ B cells exposed to HSA and the vehicle, colored by cell types. (B) Overlay of HSA and vehicle exposure on the B lymphocyte UMAP. (C) Box plots for the proportion of B cell populations that significantly changed after HSA exposure. (D, E) Scatterplots of the flow cytometry analysis showing the gating strategy to identify B and transitional B cells and boxplots showing their proportions after HSA exposure. (F) UMAP of 21,819 patients’ myeloid cells exposed to HSA and the vehicle, colored by cell populations. (G) Overlay of HSA and vehicle exposure on the myeloid cell UMAP. (H) Box plots for the proportion of myeloid cell types that significantly changed after HSA exposure. (I) UMAP of 12,692 patients’ CD4+ T cells exposed to HSA and the vehicle, colored by cell types. (J) Overlay of HSA and vehicle exposure on CD4+ T cell UMAP. (K) Box plots for the proportion of CD4+ T cells that significantly changed after HSA exposure. Fig. 3A–C and F–K have been designed using scRNA-seq in PBMCs from nine patients with AD cirrhosis. (D) and (E) were designed based on results obtained by flow cytometry in PBMCs from 10 age-matched HV. Significant differences between groups were assessed using paired t tests. AD, acutely decompensated; HSA, human serum albumin; HV, healthy volunteers; PBMC, peripheral blood mononuclear cell; pDC, plasmacytoid dendritic cell; scRNA-seq, single-cell RNA sequencing; UMAP, Uniform Manifold Approximation and Projection.
In addition to B cells, we also assessed 21,677 myeloid cells, which clustered into 13 different cell populations of monocytes (n = 7), dendritic cells (n = 3), and myeloid-derived suppressor cells and granulocyte-monocyte progenitors (n = 3) (Fig. 3F). Fig. 3G and H shows that HSA increased the frequency of monocytes with an HAVCR2+ intermediate transcriptional signature and plasmacytoid dendritic cells. The HSA-induced shift in the HAVCR2+ intermediate monocyte population was unlikely attributed to the upregulation of surface proteins resulting from cell attachment, as no changes were observed in the expression of genes associated with monocyte activation (i.e. FCGR3A, CD80, CD86, ICAM1, ITGAM, CCR2, and HLA)22 (Fig. S6A). HSA did not change the frequency of other myeloid cell populations (Fig. S6B).
We also analyzed 17,467 T lymphocytes, including 12,692 CD4+ T cells, 3,483 CD8+ T cells, and 1,292 unconventional T cells. Interestingly, HSA modified the frequency distribution of the 10 populations captured within the CD4+ T cell compartment (Fig. 3I–K and Fig. S7). Specifically, HSA decreased the frequency of activated memory CD4+ T cells while increasing central memory ITGB1+ CD4+ T cells (Fig. 3K). Finally, no significant changes were found upon HSA exposure within the 13 populations of CD8+ T cells and unconventional T cells (Fig. S8). Together, these observations confirmed at the single-cell level the ability of HSA to reprogram the transcriptional landscape of B cells and other mononuclear leukocytes isolated from patients with AD cirrhosis.
In vitro response of polymorphonuclear leukocytes (neutrophils) from patients with AD cirrhosis to HSA
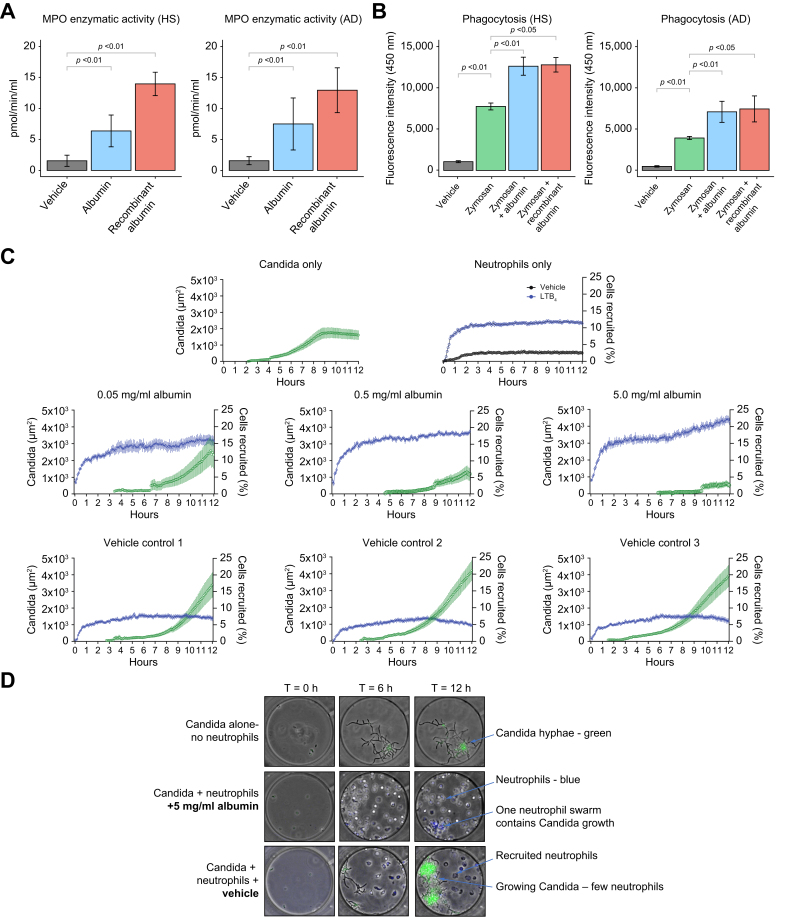
To have a more comprehensive view of the effects of HSA on blood immune cells, we designed in vitro experiments in neutrophils from patients with AD cirrhosis and HV. Given that neutrophils express fewer genes than any other leukocyte and that their lower RNA content complicates high-throughput transcriptomic analysis,23 we tested the effects of HSA on neutrophil function, including degranulation, phagocytosis, chemotaxis, and swarming. As expected, neutrophils from patients with AD cirrhosis showed impaired antimicrobial functions, including impaired phagocytosis, which is consistent with previous studies,9 as well as reduced neutrophil chemotaxis and swarming (Fig. S9). Incubation of neutrophils from both patients with AD cirrhosis and HV with either HSA or recombinant human albumin increased degranulation, as assessed by myeloperoxidase (MPO) activity in the supernatant (Fig. 4A), an essential antimicrobial protein localized mainly to the azurophil or primary neutrophil granules.24 Incubation of neutrophils from both patients with AD cirrhosis and HV with either HSA or recombinant human albumin also increased phagocytosis, as monitored by the ingestion of fluorescent-labeled zymosan particles by neutrophils (Fig. 4B). The phagocytosis findings were confirmed using fluorescent-labeled Escherichia coli (Fig. S10). We finally tested the effect of HSA on neutrophil swarming, a defensive process in which neutrophils undergo phases of highly directed and coordinated migration, followed by accumulation at sites of infection that culminate in the containment and killing of the intruding pathogen. Here, we used microfluidic devices that enable monitoring at single-cell resolution the neutrophil interactions with Candida through morphology changes from yeast to hyphae and hyphae growth.18 We also tested the effect of HSA on neutrophil phagocytosis of live Candida. Neutrophils from patients displayed severe defects in their ability to control fungal growth, as measured by the time it took for Candida hyphae to escape the neutrophil swarm and the higher total area covered by fungal growth (Fig. S9). The addition of HSA improved the ability of neutrophils to restrict fungal growth in a concentration-dependent manner (higher albumin leading to greater restriction) (Fig. 4C). Imaging showed that in the presence of HSA, neutrophils swarmed around any Candida hyphae clusters, phagocytosing them and delaying their growth (Fig. 4D and Supplementary video). In vehicle controls, neutrophils delayed the growth of Candida but were not efficient enough against clusters of Candida hyphae. These findings reveal the ability of HSA to improve the defensive functions of neutrophils isolated from immunocompromised patients with AD cirrhosis.
Fig. 4.
Effects of HSA on the host defense function of neutrophils from patients with AD cirrhosis.
(A) Neutrophils were incubated with cell medium (vehicle), HSA, or recombinant human albumin (both at 15 mg/ml) for 2 h at 37 °C in a 5% CO2 incubator. Neutrophil degranulation was assessed by measuring the MPO enzymatic activity in the cell supernatants. (B) Phagocytic capacity assessed by incubating neutrophils with FITC-conjugated zymosan bioparticles alone or in the presence of HSA and recombinant albumin for 60 min and compared with vehicle control. (C) Quantification of Candida albicans growth and neutrophil recruitment in the microfluidic device. The size of Candida hyphae clusters was quantified based on the area of green fluorescence and is shown in green. The number of neutrophils recruited was quantified and shown in blue. For the neutrophils-only condition, we compared the neutrophils entering the chambers in the presence of the vehicle (black) with the number of neutrophils in the presence of LTB4 chemoattractant (100 nM; blue). Twelve chambers per condition were quantified, and three of them are shown. (D) Microscopy images of neutrophil–Candida albicans interactions in microfluidic chambers. A similar number of Candida yeast was loaded in each chamber (time T-0). Neutrophils were loaded outside the chambers. Neutrophils migrated to the chambers, attracted by Candida-released molecules. In the presence of HSA, neutrophils actively phagocytosed Candida. Neutrophils swarmed around any Candida hyphae clusters, delaying their growth. In vehicle controls, neutrophils delayed the growth of Candida but were not efficient enough to contain clusters of Candida hyphae. (A)–(D) were designed using functional assays in freshly isolated peripheral neutrophils from six patients with AD cirrhosis and five age-matched HV. Significant differences between groups were assessed using paired t tests. AD, acutely decompensated; HSA, human serum albumin; HV, healthy volunteers; LTB4, leukotriene B4; MPO, myeloperoxidase.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.jhepr.2024.101184
The following is/are the supplementary data related to this article:
Translational study in patients with AD cirrhosis receiving HSA as therapy
The study included 49 patients with AD cirrhosis at imminent risk of ACLF at entry (T1) who developed ACLF during the index hospitalization (T2) (Fig. S11A). Of the 49 patients, 30 received HSA between T1 and T2 (albumin group), and 19 remained free of HSA (non-albumin group). We assumed that investigating patients exhibiting the most severe forms of AD cirrhosis with the use of longitudinal data would limit the effect of interindividual variability on the assessment of HSA effects. The clinical characteristics and laboratory data of all patients at T1 and T2 are given in Table S3. Characteristics were similar in terms of standard laboratory values, including clinical differential blood counts for neutrophils, monocytes, and lymphocytes (Fig. S12A), which strongly correlated with RNA-seq-inferred blood counts (Fig. S12B). As expected, patients presented organ failures, higher white blood cell counts, and elevated levels of inflammatory markers at T2 compared with HV (Tables S3 and S4).
We next considered characteristics at T1 and T2 in patients in the albumin and non-albumin groups. In both groups, progression to ACLF from T1 to T2 was associated with significant increases in the model for end-stage liver disease (MELD) score. However, except for serum creatinine, which increased at T2, there were no significant within-group changes regarding the rest of longitudinally collected standard laboratory data and circulating levels of inflammatory mediators (Table 1). However, there were some in between-group differences. The delay between T1 and T2 was shorter in the albumin group than in the non-albumin group. Compared with patients in the non-albumin group, those in the albumin group had significantly higher CRP, IL-6, and IL-10 levels at T1 and higher Chronic Liver Failure Consortium organ failure (CLIF-C OF) score and creatinine, CRP, IL-6, and MCP-1 levels at T2. Moreover, a higher percentage of patients in the albumin group died by 28 and 90 days. The difference in survival was expected considering the higher prevalence of SBP and HRS-AKI in the albumin group.
Table 1.
Characteristics of patients at time 1 (T1) and time 2 (T2) and deaths in the albumin and non-albumin groups.
| Characteristics | Albumin group (n = 30) |
Non-albumin group (n = 19) |
p value |
|||||
|---|---|---|---|---|---|---|---|---|
| T1 | T2 | T1 | T2 | T2 vs. T1 (albumin group) |
T2 vs. T1 (non-albumin group) |
T1 (between groups) |
T2 (between groups) |
|
| Markers of organ function | ||||||||
| MELD score, mean ± SD | 19.2 ± 5.3 | 27.0 ± 6.6 | 19.0 ± 5.7 | 24.0 ± 7.5 | <0.01 | 0.01 | 0.92 | 0.17 |
| CLIF-C OF score, mean ± SD | 7.0 ± 1.0 | 10.5 ± 2.6 | 7.4 ± 1.3 | 8.9 ± 1.5 | <0.01 | <0.01 | 0.26 | 0.01 |
| Organ system failure, n (%) | ||||||||
| Liver failure | 6 (20) | 7 (23) | 4 (21) | 6 (32) | 1 | 0.71 | 1 | 0.76 |
| Kidney failure | 0 (0) | 23 (77) | 0 (0) | 13 (68) | <0.01 | <0.01 | 1 | 0.76 |
| Circulatory failure | 0 (0) | 10 (33) | 1 (5) | 1 (5) | <0.01 | 1 | 1 | 0.69 |
| Cerebral failure | 0 (0) | 6 (20) | 0 (0) | 0 (0) | <0.01 | 1 | 0.82 | 0.31 |
| Coagulation failure | 0 (0) | 4 (13) | 0 (0) | 4 (22) | 0.12 | 0.10 | 1 | 0.01 |
| Respiratory failure | 0 (0) | 9 (30) | 0 (0) | 0 (0) | <0.01 | 1 | 1 | 0.02 |
| ACLF grade, no. (%) | ||||||||
| 1 | 0 (0) | 13 (43) | 0 (0) | 13 (72) | <0.01 | <0.01 | 1 | 0.1 |
| 2–3 |
0 (0) |
17 (57) |
0 (0) |
5 (28) |
<0.01 |
0.05 |
1 |
0.1 |
| Precipitating events, n (%) | ||||||||
| Infection as precipitant at T1 or T2 | 14 (47) | 20 (67) | 5 (26) | 7 (37) | 0.19 | 0.73 | 0.5 | 0.03 |
| Alcohol-related hepatitis as precipitant at T1 or T2 |
14 (50) |
14 (47) |
7 (39) |
7 (37) |
1 |
1 |
0.4 |
0.21 |
| Laboratory data | ||||||||
| International normalized ratio, median (IQR) | 1.6 (1.4–1.7) | 1.6 (1.5–2.0) | 1.5 (1.2–1.8) | 1.4 (1.3–2.2) | 0.08 | 0.48 | 0.7 | 0.27 |
| Total bilirubin (mg/L), median (IQR) | 3.7 (2.0–9.0) | 3.6 (2.4–11.3) | 3.2 (1.3–9.5) | 5.0 (1.0–13.5) | 0.83 | 0.99 | 0.72 | 0.52 |
| Serum creatinine (mg/dl), median (IQR) | 1.2 (1.0–1.6) | 2.7 (1.9–3.4) | 1.4 (1.1–1.6) | 2.1 (1.4–2.4) | <0.01 | 0.01 | 0.59 | 0.02 |
| Serum sodium (mmol/L), mean ± SD | 131 ± 5.8 | 133 ± -8.4 | 134.0 ± 6.9 | 132 ± 6.9 | 0.31 | 0.53 | 0.18 | 0.74 |
| Serum albumin (g/dl), median (IQR) | 2.6 (2.2–3.2) | 3.0 (2.3–3.5) | 2.7 (2.5–3.4) | 3.0 (2.8–3.1) | 0.21 | 0.36 | 0.23 | 0.89 |
| White cell count ( × 103/mm3), median (IQR) | 8.3 (7.0–10.3) | 11.9 (6.2–13.7) | 6.6 (4.6–8.6) | 8.1 (5.8–12.4) | 0.06 | 0.12 | 0.14 | 0.31 |
| Absolute lymphocyte count ( × 103/mm3), median (IQR) | 0.9 (0.6–1.3) | 1.1 (0.7–1.6) | 1.1 (0.8–1.5) | 1.2 (0.8–1.8) | 0.25 | 0.54 | 0.23 | 0.48 |
| Absolute monocyte count ( × 103/mm3), median (IQR) | 0.8 (0.6–1.1) | 1.0 (0.6–1.3) | 0.6 (0.3–1.0) | 0.7 (0.6–0.9) | 0.1 | 0.48 | 0.28 | 0.08 |
| Absolute neutrophil count ( × 103/mm3), median (IQR) | 6.1 (4.2–7.7) | 8.9 (4.2–11.8) | 4.0 (3.4–6.4) | 4.5 (3.0–7.8) | 0.12 | 0.73 | 0.15 | 0.05 |
| C-reactive protein (mg/L), median (IQR) |
31.3 (19.7–54.0) |
33.5 (19.4–90.4) |
13.3 (7.4–25.0) |
16.1 (11.4–21.8) |
0.5 |
0.69 |
<0.01 |
0.01 |
| Blood levels of protein mediators of inflammation (pg/ml), median (IQR) | ||||||||
| Eotaxin | 66.0 (43.9–97.1) | 86.3 (58.6–104.1) | 71.0 (43.7–121.0) | 64.3 (50.1–121.9) | 0.1 | 1 | 0.47 | 0.53 |
| Granulocyte-colony stimulating factor | 18.8 (4.4–43.2) | 22.5 (9.0–73.8) | 23.9 (7.6–99.6) | 16.9 (3.6–70.9) | 0.59 | 0.75 | 0.63 | 0.61 |
| IFN-α2 | 10.0 (2.2–24.8) | 16.8 (2.4–33.4) | 12.8 (7.6–22.0) | 17.6 (8.2–22.7) | 0.43 | 0.5 | 0.68 | 0.97 |
| IFN-γ | 33.3 (7.0–86.0) | 23.8 (12.1–97.2) | 19.2 (7.2–98.7) | 25.9 (7.9–45.8) | 0.93 | 0.84 | 0.76 | 0.57 |
| IL-1α | 4.3 (1.7–7.2) | 2.0 (0.8–6.1) | 2.5 (1.1–3.3) | 2.0 (0.3–3.9) | 0.3 | 0.71 | 0.12 | 0.45 |
| IL-1β | 5.4 (3.1–10.8) | 4.7 (2.3–13.0) | 5.2 (1.8–9.7) | 5.2 (1.9–8.7) | 0.93 | 0.89 | 0.71 | 0.64 |
| IL-6 | 24.7 (13.6–46.5) | 52.5 (24.3–168.9) | 9.3 (6.8–18.1) | 16.9 (8.8–23.1) | 0.16 | 0.1 | <0.01 | <0.01 |
| IL-8 | 5.5 (3.5–10.7) | 12.6 (4.0–18.1) | 4.3 (2.6–13.0) | 4.3 (1.0–9.0) | 0.19 | 0.6 | 0.44 | 0.03 |
| IL-10 | 10.0 (3.9–30.4) | 13.1 (3.8–26.4) | 2.4 (1.2–3.6) | 4.7 (3.2–5.8) | 0.97 | 0.08 | 0.03 | 0.15 |
| Interferon-inducible protein 10 | 231 (123–374) | 231 (169–615) | 214 (166–482) | 237 (141–487) | 0.34 | 1 | 0.69 | 0.71 |
| Monocyte chemoattractant protein 1 | 179 (147–257) | 349 (230–533) | 176 (158–279) | 198.5 (100–281) | <0.01 | 0.8 | 0.74 | <0.01 |
| Macrophage inflammatory protein 1α | 11.6 (8.4–23.5) | 15.2 (9–28.9) | 8.5 (2–18) | 13.3 (4.5–19.4) | 0.38 | 0.41 | 0.21 | 0.26 |
| Macrophage inflammatory protein 1β | 15.7 (11.7–19.7) | 19.9 (13.8–30.6) | 15.9 (12–22.7) | 15.2 (11.7–19.6) | 0.08 | 0.7 | 0.56 | 0.15 |
| Tumor necrosis factor | 30.8 (18.1–42) | 35.3 (24–66.6) | 37.6 (18.8–47.2) | 35.6 (20.8–49.3) | 0.31 | 0.89 | 0.59 | 0.62 |
| Vascular endothelial growth factor | 4.3 (0.8–8.2) | 6.3 (2.6–16.4) | 5.2 (1.9–12.7) | 7.6 (4.7–11.7) | 0.17 | 0.25 | 0.57 | 0.53 |
Normal values for blood inflammatory mediators are shown in Table S4. There was no significant difference between the albumin and non-albumin groups in terms of age (mean ± SD, 61.0 ± 7.9 and 59.1 ± 10.8 years, respectively) and number (%) of males (21 [70] and 12 [63], respectively). A higher percentage of patients in the albumin group than in the no-albumin group died by 28 days (30% [9/30] and 0%, respectively) and 90 days (67% [20/30] and 16% [3/19], respectively).
ACLF, acute-on-chronic liver failure; CLIF-C OF, Chronic Liver Failure Consortium organ failure; MELD, model for end-stage liver disease.
Transcriptional characteristics of all patients at T1 and T2
Analyzing differentially expressed genes (DEGs) in two comparisons, T1 vs. HV and T2 vs. HV, we observed, as expected, strong similarities between the two disease stages. Thus, the number of DEGs was high in the two comparisons; that is, there were 4,615 DEGs in T1 relative to HV and 4,529 in T2 relative to HV. Of the 4,615 DEGs associated with T1, 3,929 (85%) overlapped with the DEGs assigned to T2 (Fig. S11B, top). By comparing effect-size changes in T1 vs. HV with T2 vs. HV, we observed a highly concordant magnitude of changes between the two signatures (Fig. S11B, bottom). Next, we used Quantitative Set Analysis for Gene Expression (QuSAGE) to analyze the differential expression of blood transcription modules (BTMs). Among the 258 annotated BTMs, the total number of DE BTMs (false discovery rate [FDR] <0.05) was 190 in T1 vs. HV and 186 in T2 vs. HV (Fig. S11C, top; Table S5A). These findings indicated extensive changes in the blood transcription module space in both T1 and T2. Strikingly, only 20 BTMs had specific differential expression relative to HV, in either T1 (12 modules) or T2 (eight modules), whereas 179 DE BTMs were shared with a concordant sign, of which 108 were upregulated and 71 downregulated (Table S5A). Fig. S11C (bottom) shows the 20 top gene modules for shared upregulated BTMs and 20 top gene modules for shared downregulated BTMs. Shared upregulated BTMs were related to innate immunity, including those related to Toll-like receptors and inflammatory signaling, interferon-alpha response, and innate immune cells (i.e. neutrophils, monocytes, and dendritic cells). Shared downregulated BTMs were related to T and NK cells and antigen presentation. These results were consistent with those obtained when analyzing gene signatures for the 10 main immune cell types (Fig. S13A) and 29 fine immune cell types (Fig. S13B). Collectively, our analyses of bulk blood RNA-seq data provided consistent results that highlighted the similarity of the transcriptional landscape in T1 and T2, both disease’s stages being characterized by simultaneity between increases in gene signatures related to innate immunity and decreases in gene signatures related to adaptive immunity.
Whole blood gene signatures associated with HSA treatment
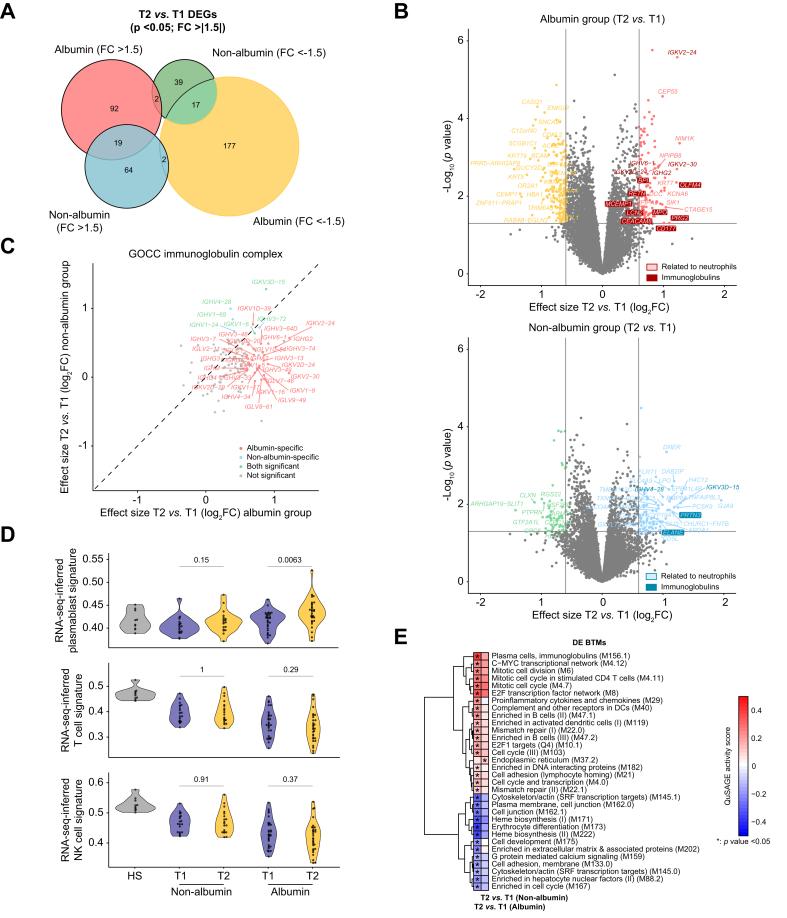
We compared differential gene expression between T2 and T1 for the albumin and non-albumin groups and found that gene signatures differed between the two groups. Indeed, 269 DEGs (92 upregulated, 177 downregulated) were specific for the albumin group, whereas 103 DEGs (64 upregulated, 39 downregulated) were specific for the non-albumin group; only 36 DEGs with a concordant sign were shared by the two groups (Fig. 5A). Identification of DEGs using volcano plots (Fig. 5B) illustrated the group specificity of both upregulated and downregulated genes. A broad variety of immunoglobulin genes (see below) and several major neutrophil genes (CD177, OLFM4, PRG2, MPO, BPI, RETN, LCN2, CEACAM8, MCEMP1) were upregulated in the albumin group (Fig. 5B, top), but not in the non-albumin group (Fig. 5B, bottom). These findings agree with the observed in vitro responses of B cells and neutrophils to albumin. Because analysis of DEGs drew our attention to immunoglobulin genes, we compared the effect-size changes between T2 and T1 within each group for each of the 125 genes (including 120 immunoglobulin genes) that are included in the Gene Ontology gene set labeled ‘GOCC Immunoglobulin Complex’. We found that effect-size changes were greater in the albumin group (33 upregulated immunoglobulin genes) than in the non-albumin group (only 6) (Fig. 5C). Gene coding for constant regions of immunoglobulin heavy chains (IGHM, IGHG2, IGHG3, IGHG4, and IGHA2) were specifically upregulated in the albumin group. These findings are consistent with the results of SingleR analysis, which showed increases in gene signatures for plasmablasts that were specific for the albumin group (Fig. 5D, top). Immunoglobulin genes that were specifically upregulated in the albumin group also included genes for constant regions of immunoglobulin light chains kappa (IGKC) and lambda (IGLC1), genes for the V regions of the variable domain of immunoglobulin heavy chains, and genes for the V regions of the variable domain of light chains kappa and lambda. The three genes that were specifically upregulated in the non-albumin group were genes coding for the V regions of the variable domain of immunoglobulin heavy chains (Fig. 5C). Together these findings indicate an extensive upregulation of gene coding for immunoglobulins that were specific for patients who had received HSA.
Fig. 5.
Whole blood gene signatures associated with HSA treatment.
(A) Euler plot showing DEGs between T2 and T1 for the albumin and non-albumin groups. DEGs were defined by an absolute FC greater than 1.5 and p <0.05. (B) Volcano plots showing differential expression effect size (log2 FC) between T2 and T1 plotted against significance (-log10p) for the albumin (top) and non-albumin (bottom) groups. In both volcano plots, gray points indicate genes with no significant difference in expression between T2 and T1 (with absolute FC <1.5 and p >0.05, i.e. -log10p <1.3). Colored points indicate DEGs, either upregulated or downregulated. The neutrophil-related genes are highlighted. (C) Differential expression effect size (log2 FC) between T2 and T1 in the albumin group compared with corresponding differential effect size in the non-albumin group. Only genes included in the Gene Ontology gene set ‘GOCC_Immunoglobulin Complex’ are shown here. DEGs with a concordant sign were considered as shared. (D) Violin plots of RNA-seq inferred signatures at T1 and T2 for plasmablasts, NK cells, and T cells, determined using the SingleR software, in the albumin and non-albumin groups. Baseline values in HV are also shown. Values of p are from Kruskal–Wallis tests. (E) Heat map of 32 DE BTMs between T2 and T1 that were specific for either the albumin or non-albumin group. Thirty-one DE BTMs were specific for the albumin group, whereas only one DE BTM (‘endoplasmic reticulum [M37.2]’) was specific for the non-albumin group. BTMs were hierarchically clustered based on QuSAGE activity scores obtained in the albumin group. Asterisks denote p <0.05 obtained when comparing probability density functions with QuSAGE. Color represents QuSAGE activity score. BTM, blood transcription module; DE, differentially expressed; DEG, differentially expressed gene; FC, fold change; HSA, human serum albumin; NK, natural killer; QuSAGE, Quantitative Set Analysis for Gene Expression; RNA-seq, RNA sequencing; T1, time 1; T2, time 2.
Next, we applied QuSAGE to identify DE BTMs between T2 and T1 within each group. We first observed that the number of DE BTMs was greater in the albumin group than in the non-albumin group (36 vs. 6, respectively; Table S5B). Only one DE BTM, related to endoplasmic reticulum, was specifically upregulated in the non-albumin group (Fig. 5E). In sharp contrast, 31 DE BTMs (13 downregulated, 18 upregulated) were specific for the albumin group (Fig. 5E). Downregulated modules were related to erythropoiesis, cytoskeleton, or cell junction (Fig. 5E), whereas upregulated BTMs were related to activated dendritic cells, complement and other receptors in dendritic cells, cytokines, and chemokines (Fig. 5E). Upregulated BTMs specific for the albumin group also comprised modules related to B cells (including those related to enriched in B cells, plasma cells, and immunoglobulins), mismatch repair, cell cycle, and mitosis. Of note, upregulation of BTMs related to B cells in the albumin group was confirmed after adjusting the transcriptomics data by disease severity, as estimated by the MELD score (Fig. S14). Moreover, a direct and significant positive correlation was observed between the mean daily HSA dose administered to the patients and the expansion of the B cell compartment (Fig. S15). However, no evidence for HSA induction of NF-κB, a transcription factor involved in B-cell development and survival,25,26 was observed in our study (Fig. S16). Of note, RNA-seq-inferred signatures for T and NK cells (Fig. 5D, middle and bottom panels; and Fig. S17A and B) as well as gene modules related to these cells (Table S5B), whose downregulation is a hallmark of patients with AD cirrhosis without and with ACLF (Fig. S11C), remained all downregulated among patients treated with HSA. Nevertheless, we observed that a BTM labeled ‘mitotic cell cycle in stimulated CD4+ T cells (M4.11)’ was specifically upregulated among patients treated with HSA (Fig. 5E), which suggests some activation of transcription in CD4+ T cells. Consistent with this, we found no overlap between member genes of the M4.11 BTM and genes used for SingleR analysis of CD4+ T cells (Fig. S18). These findings were validated by scRNA-seq, in which the gene signature score for this BTM across all CD4+ T cells computed using the Ucell package27 showed a significantly higher cell density in HSA-treated cells relative to the vehicle (Fig. S19).
Discussion
The results of the current investigation contribute to understanding the mechanisms by which albumin administration is associated with a lower rate of infections and better survival in patients with AD cirrhosis. The investigation was performed in vitro in peripheral leukocytes and in vivo in patients with AD cirrhosis receiving albumin therapy. The investigation used cutting-edge technologies (i.e. bulk and scRNA-seq and swarming assays in microfluidic arenas) and gold-standard functional assays to assess leukocyte antimicrobial functions. The results of this investigation revealed that albumin modulates the peripheral immune system by attenuating B-cell depletion and revitalizing neutrophil functions in patients with AD cirrhosis.
To our knowledge, this is the first study to use longitudinal transcriptomics to characterize the effects of albumin on immune cells in patients with AD cirrhosis. The design of the study, particularly in those aspects related to the selection of patients, was performed after considering that patients hospitalized with ACLF, who are known to present an extremely dynamic clinical course with marked changes in the magnitude of systemic inflammation,2,17 were clearly poor candidates for inclusion. Other phenotypes, including ‘unstable’ and ‘stable’ AD cirrhosis, were also not considered because they are associated with moderate systemic inflammation, which may further decrease during hospitalization, and we were interested in including only patients with intense systemic inflammation to better assess immune changes induced by albumin. For this reason, we chose patients with AD cirrhosis with imminent risk of developing ACLF, in whom the magnitude of systemic inflammation was similar to that seen in ACLF. We included only patients who received albumin for well-established indications (paracentesis, prevention of HRS-AKI associated with SBP, or HRS-AKI treatment)12 even though this would introduce the bias of higher severity in patients receiving albumin. We did not select patients receiving albumin within 1 month before T1, those with a too-long interval between the two assessments of bulk blood RNA-seq, and those with a delay of >10 days between the last dose of albumin and T2. Finally, although albumin dosage and duration of treatment varied according to indications, we decided to analyze data in all patients irrespective of albumin dosage. The similarity of transcriptional characteristics as well as most clinical characteristics, standard laboratory values, and levels of cytokines in patients with AD cirrhosis before and after developing ACLF confirmed our assumption that patients included in the study were in a relatively steady state of systemic inflammation during the study.
The major finding of the scRNA-seq and bulk RNA-seq in blood cells from patients with AD cirrhosis was that albumin administration triggered signals that caused an expansion of the B cell compartments, and likely the CD4+ T cell compartments, while activating some mononuclear myeloid cells. These findings are extremely relevant, considering that the prevalence of bacterial infections at admission in patients with AD cirrhosis is very high (37.3 and 25.1% in patients with and without ACLF, respectively).10,28 Among the uninfected patients at admission, 46% with and 18% without ACLF develop bacterial infection during hospitalization. The existence of a profound impairment of the innate immune system, which is highlighted by impaired neutrophil and monocyte antimicrobial functions and lymphocyte depletion, likely explains such a high risk of infections in these patients.29 Of note, because changes in immune-cell transcriptome seen after albumin administration occurred in the absence of changes in albuminemia, the effects of albumin on the transcriptome of immune cells cannot be merely explained by correction of hypoalbuminemia but rather by qualitative differences between the ‘commercial’ albumin and the circulating endogenous albumin in patients with AD cirrhosis, which is known to be post-translationally modified and highly oxidized.30
Some mechanisms could explain the effect of albumin on immune cells. Immune cells can be activated by osmotic stress.31 Therefore, the movement of water from cells to the extracellular milieu (osmotic stress) could be a mechanism by which albumin exerted immunomodulatory effects in our in vitro cell experiments. However, we did not see the same response in PBMCs incubated with mannitol, a compound sharing the iso-oncotic properties of albumin. Another mechanism could be the induction of NF-κB, a master transcription factor in B cells25 that has been reported to be activated by albumin in a human renal proximal tubule-derived cell line.26 However, in our study, we did not find any sign of NF-κB induction in response to albumin. Because extracellular acidification is known to reprogram immune cell responsiveness,32 another possibility is that a change in pH caused by albumin, which is a highly soluble acidic molecule, could account for the activation of immune cells. This can also occur at the intracellular level because albumin is rapidly internalized by mononuclear myeloid cells16 and intracellular acidification results in changes in immune cell performance.33 In contrast, the interaction of albumin with FcRn or with voltage-gated proton channels, which leads to stimulation of phagocytosis and degranulation,19,34,35 respectively, could mechanistically explain the activation of neutrophil antimicrobial function by albumin. In this regard, we observed that the immunostimulatory actions of albumin in mononuclear leukocytes were partially inhibited by an antibody blocking FcRn.
This study has some limitations. First, there is a difference in severity between the two study groups. Second, our results are mostly descriptive, which makes it difficult to directly prove immune restoration by albumin. The third limitation is related to the study design. Indeed, we leveraged the prespecified collection of blood for RNA-seq in patients of the PREDICT study at admission (when they did not have ACLF) and at the time of ACLF development to identify patients who did or did not receive albumin during the progression to ACLF. Thus, we do not report the results of a controlled study specifically designed to investigate the effects of prespecified systematic albumin administration, for example, on the development of ACLF. The ideal protocol would consist in daily administration of albumin with the objective of, for example, maintaining blood levels of albumin above 3.0 g/dl, as in the ATTIRE trial.36 Nevertheless, it is noteworthy that conducting a prospective randomized controlled trial of albumin for preventing ACLF in patients who present with pre-ACLF would face major methodological issues. Indeed, there are currently no criteria to identify patients with pre-ACLF among those who present for AD cirrhosis.17 Therefore, performing randomized controlled trials that enroll well-matched patients with AD cirrhosis is a big challenge, and criteria for stratifying patients according to the risk of ACLF is an unmet medical need.
In conclusion, the results of this study suggest that albumin promotes the expansion of the B cell compartment and improves neutrophil antimicrobial functions. These findings contribute to understanding the clinical benefits of albumin therapy such as reducing the incidence rate of infections in patients with AD cirrhosis.13
Abbreviations
ACLF, acute-on-chronic liver failure; AD, acutely decompensated; BTM, blood transcription module; DE, differentially expressed; DEG, differentially expressed genes; FC, fold change; FcRn, neonatal Fc receptor; FDR, false discovery rate; GSEA, gene set enrichment analysis; HRS-AKI, hepatorenal syndrome–acute kidney injury; HSA, human serum albumin; HV, healthy volunteers; MELD, model for end-stage liver disease; MPO, myeloperoxidase; NES, normalized enrichment score; NK, natural killer; PBMC, peripheral blood mononuclear cell; QuSAGE, Quantitative Set Analysis for Gene Expression; RNA-seq, RNA sequencing; SBP, spontaneous bacterial peritonitis; scRNA-seq, single-cell RNA sequencing; UMAP, Uniform Manifold Approximation and Projection.
Financial support
This research was supported by EF CLIF, a private, non-profit organization receiving unrestricted grants from Grifols and Fundació Privada Cellex. EF CLIF is a partner and/or coordinator of several European Union (EU) Horizon 2020 program projects (Nos. 825694 and 847949). This study was also supported by Ministerio de Ciencia e Innovacion/Agencia Estatal Investigación MCIN/AEI, PID2022-138970OB-I00 10.13039/501100011033/FEDER, UE (to JC) and RTI2018-093894-B-I00 (to AC). LJ-G has held an FPU PhD fellowship (FPU19/04886) from the Spanish Ministry of Science, Innovation and Universities. EW was an EF CLIF Research Fellow who received grants from Fundació Privada Cellex and the Fondation Bettencourt-Schueller. IH received a Marie Skłodowska-Curie grant agreement (No. 75451) from the EU’s Horizon 2020 Research and Innovation Program. The funders had no influence on the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflicts of interest
MB is part of the speakers’ bureau for Grifols SA, Octapharma AG, Shire/Takeda, CLS Behring GmbH, and PPTA and is a consultant for Grifols SA, CLS Behring GmbH, Martin Pharmaceuticals, and Shire/Takeda. HH is a co-founder and shareholder of Omniscope, a scientific advisory board member of MiRXES, and a consultant to Moderna. RJ has research collaborations with Yaqrit and Takeda. He is the inventor of OPA, which has been patented by UCL and licensed to Mallinckrodt Pharma. He is also the founder of Yaqrit Ltd, a spin-out company from University College London and Thoeris Ltd. JCN is a scientific consultant of Omniscope. The remaining authors disclose no conflicts.
Please refer to the accompanying ICMJE disclosure forms for further details.
Authors’ contributions
Study concept and design: VA, RM. Acquisition of data and sample measurements: MC, CLV, BRG, XM, AV, AMA, EW, JT, JF, MB, RJ, PA. Bioinformatic and statistical analyses: JJL, IH, FA, LJG, JCN, AMA, DM. Drafting of the manuscript: JC, VA, RM. Critical revision of the manuscript for important intellectual content: EW, JC, JJL, IH, JT, FA, JF, LJG, JCN, AV, AMA, MB, RJ, PA, GM, AC, HH. Study supervision: RM, VA, HH.
Data availability statement
All data associated with this work is presented in the main manuscript or Supplementary material. For availability of any other type of data, contact the corresponding authors.
Acknowledgements
We thank Dr Anna Bosch, Dr Lidia García-Campmany, Ms Anna Curto, Ms Cristina Sanchez, and Mr Alex Amoros for their help.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2024.101184.
Contributor Information
Joan Clària, Email: jclaria@clinic.cat.
Richard Moreau, Email: richard.moreau@inserm.fr.
Supplementary data
The following are the supplementary data to this article:
References
- 1.Moreau R., Jalan R., Ginès P., et al. Acute-on-chronic liver failure is a distinct syndrome that develops in patients with acute decompensation of cirrhosis. Gastroenterology. 2013;144:1426–1437. doi: 10.1053/j.gastro.2013.02.042. [DOI] [PubMed] [Google Scholar]
- 2.Arroyo V., Moreau R., Jalan R. Acute-on-chronic liver failure. N Engl J Med. 2020;382:2137–2145. doi: 10.1056/NEJMra1914900. [DOI] [PubMed] [Google Scholar]
- 3.Weiss E., de la Grange P., Defaye M., et al. Characterization of blood immune cells in patients with decompensated cirrhosis including ACLF. Front Immunol. 2021;11 doi: 10.3389/fimmu.2020.619039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clària J., Stauber R.E., Coenraad M.J., et al. Systemic inflammation in decompensated cirrhosis: characterization and role in acute-on-chronic liver failure. Hepatology. 2016;64:1249–1264. doi: 10.1002/hep.28740. [DOI] [PubMed] [Google Scholar]
- 5.López-Vicario C., Checa A., Urdangarin A., et al. Targeted lipidomics reveals extensive changes in circulating lipid mediators in patients with acutely decompensated cirrhosis. J Hepatol. 2020;73:817–828. doi: 10.1016/j.jhep.2020.03.046. [DOI] [PubMed] [Google Scholar]
- 6.Bernsmeier C., Pop O.T., Singanayagam A., et al. Patients with acute-on-chronic liver failure have increased numbers of regulatory immune cells expressing the receptor tyrosine kinase MERTK. Gastroenterology. 2015;148:603–615. doi: 10.1053/j.gastro.2014.11.045. [DOI] [PubMed] [Google Scholar]
- 7.Korf H., du Plessis J., van Pelt J., et al. Inhibition of glutamine synthetase in monocytes from patients with acute-on-chronic liver failure resuscitates their antibacterial and inflammatory capacity. Gut. 2019;68:1872–1883. doi: 10.1136/gutjnl-2018-316888. [DOI] [PubMed] [Google Scholar]
- 8.O’Brien A.J., Fullerton J.N., Massey K.A., et al. Immunosuppression in acutely decompensated cirrhosis is mediated by prostaglandin E2. Nat Med. 2014;20:518–523. doi: 10.1038/nm.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stadlbauer V., Mookerjee R.P., Wright G.A., et al. Role of Toll-like receptors 2, 4, and 9 in mediating neutrophil dysfunction in alcoholic hepatitis. Am J Physiol Gastrointest Liver Physiol. 2009;296:G15–G22. doi: 10.1152/ajpgi.90512.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernández J., Acevedo J., Wiest R., et al. Bacterial and fungal infections in acute-on-chronic liver failure: prevalence, characteristics and impact on prognosis. Gut. 2018;67:1870–1880. doi: 10.1136/gutjnl-2017-314240. [DOI] [PubMed] [Google Scholar]
- 11.Fernández J., Piano S., Bartoletti M., et al. Management of bacterial and fungal infections in cirrhosis: the MDRO challenge. J Hepatol. 2021;75:101–117. doi: 10.1016/j.jhep.2020.11.010. [DOI] [PubMed] [Google Scholar]
- 12.European Association for the Study of the Liver EASL Clinical Practice Guidelines for the management of patients with decompensated cirrhosis. J Hepatol. 2018;69:406–460. doi: 10.1016/j.jhep.2018.03.024. [DOI] [PubMed] [Google Scholar]
- 13.Caraceni P., Riggio O., Angeli P., et al. Long-term albumin administration in decompensated cirrhosis: an open-label randomised trial. Lancet. 2018;391:2417–2429. doi: 10.1016/S0140-6736(18)30840-7. [DOI] [PubMed] [Google Scholar]
- 14.Fernández J., Clària J., Amorós A., et al. Effects of albumin treatment on systemic and portal hemodynamics and systemic inflammation in patients with decompensated cirrhosis. Gastroenterology. 2019;157:149–162. doi: 10.1053/j.gastro.2019.03.021. [DOI] [PubMed] [Google Scholar]
- 15.Bihari S., Bannard-Smith J., Bellomo R. Albumin as a drug: its biological effects beyond volume expansion. Crit Care Resusc. 2020;22:257–265. doi: 10.1016/S1441-2772(23)00394-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casulleras M., Flores-Costa R., Duran-Güell M., et al. Albumin internalizes and inhibits endosomal TLR signaling in leukocytes from patients with decompensated cirrhosis. Sci Transl Med. 2020;12 doi: 10.1126/scitranslmed.aax5135. [DOI] [PubMed] [Google Scholar]
- 17.Trebicka J., Fernandez J., Papp M., et al. The PREDICT study uncovers three clinical courses of acutely decompensated cirrhosis that have distinct pathophysiology. J Hepatol. 2020;73:842–854. doi: 10.1016/j.jhep.2020.06.013. [DOI] [PubMed] [Google Scholar]
- 18.Ellett F., Jalali F., Marand A.L., et al. Microfluidic arenas for war games between neutrophils and microbes. Lab Chip. 2019;19:1205–1216. doi: 10.1039/c8lc01263f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pyzik M., Rath T., Lencer W.I., et al. FcRn: the architect behind the immune and nonimmune functions of IgG and albumin. J Immunol. 2015;194:4595–4603. doi: 10.4049/jimmunol.1403014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart A., Ng J.C., Wallis G., et al. Single-cell transcriptomic analyses define distinct peripheral B cell subsets and discrete development pathways. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.602539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polet H., Spieker-Polet H. Serum albumin is essential for in vitro growth of activated human lymphocytes. J Exp Med. 1975;142:949–959. doi: 10.1084/jem.142.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapellos T.S., Bonaguro L., Gemünd I., et al. Human monocyte subsets and phenotypes in major chronic inflammatory diseases. Front Immunol. 2019;10:2035. doi: 10.3389/fimmu.2019.02035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grieshaber-Bouyer R., Radtke F.A., Cunin P., et al. The neutrotime transcriptional signature defines a single continuum of neutrophils across biological compartments. Nat Commun. 2021;12:2856. doi: 10.1038/s41467-021-22973-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amulic B., Cazalet G.L., Hayes K.D., et al. Neutrophil function: from mechanisms to disease. Annu Rev Immunol. 2012;30:459–489. doi: 10.1146/annurev-immunol-020711-074942. [DOI] [PubMed] [Google Scholar]
- 25.Sasaki Y., Iwai K. Roles of the NF-κB pathway in B-lymphocyte biology. Curr Top Microbiol Immunol. 2016;393:177–209. doi: 10.1007/82_2015_479. [DOI] [PubMed] [Google Scholar]
- 26.Drumm K., Bauer B., Freudinger R., et al. Albumin induces NF-κB expression in human proximal tubule-derived cells (IHKE-1) Cell Physiol Biochem. 2002;12:187–196. doi: 10.1159/000066278. [DOI] [PubMed] [Google Scholar]
- 27.Andreatta M., Carmona S.J. UCell: robust and scalable single-cell gene signature scoring. Comput Struct Biotechnol J. 2021;19:3796–3798. doi: 10.1016/j.csbj.2021.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trebicka J., Fernandez J., Papp M., et al. PREDICT identifies precipitating events associated with the clinical course of acutely decompensated cirrhosis. J Hepatol. 2021;74:1097–1108. doi: 10.1016/j.jhep.2020.11.019. [DOI] [PubMed] [Google Scholar]
- 29.Arroyo V., Moreau R., Kamath P.S., et al. Acute-on-chronic liver failure in cirrhosis. Nat Rev Dis Primers. 2016;2 doi: 10.1038/nrdp.2016.41. [DOI] [PubMed] [Google Scholar]
- 30.Alcaraz-Quiles J., Casulleras M., Oettl K., et al. Oxidized albumin triggers a cytokine storm in leukocytes through P38 mitogen-activated protein kinase: role in systemic inflammation in decompensated cirrhosis. Hepatology. 2018;68:1937–1952. doi: 10.1002/hep.30135. [DOI] [PubMed] [Google Scholar]
- 31.Ip W.K., Medzhitov R. Macrophages monitor tissue osmolarity and induce inflammatory response through NLRP3 and NLRC4 inflammasome activation. Nat Commun. 2015;6:6931. doi: 10.1038/ncomms7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang W., Le J., Wang P.Y., et al. Extracellular acidity reprograms macrophage metabolism and innate responsiveness. J Immunol. 2021;206:3021–3031. doi: 10.4049/jimmunol.2100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murase M., Kawasaki T., Hakozaki R., et al. Intravesicular acidification regulates lipopolysaccharide inflammation and tolerance through TLR4 trafficking. J Immunol. 2018;200:2798–2808. doi: 10.4049/jimmunol.1701390. [DOI] [PubMed] [Google Scholar]
- 34.Vidarsson G., Stemerding A.M., Stapleton N.M., et al. FcRn: an IgG receptor on phagocytes with a novel role in phagocytosis. Blood. 2006;108:3573–3579. doi: 10.1182/blood-2006-05-024539. [DOI] [PubMed] [Google Scholar]
- 35.Zhao R., Dai H., Arias R.J., et al. Direct activation of the proton channel by albumin leads to human sperm capacitation and sustained release of inflammatory mediators by neutrophils. Nat Commun. 2021;12:3855. doi: 10.1038/s41467-021-24145-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.China L., Freemantle N., Forrest E., et al. A randomized trial of albumin infusions in hospitalized patients with cirrhosis. N Engl J Med. 2021;384:808–817. doi: 10.1056/NEJMoa2022166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this work is presented in the main manuscript or Supplementary material. For availability of any other type of data, contact the corresponding authors.