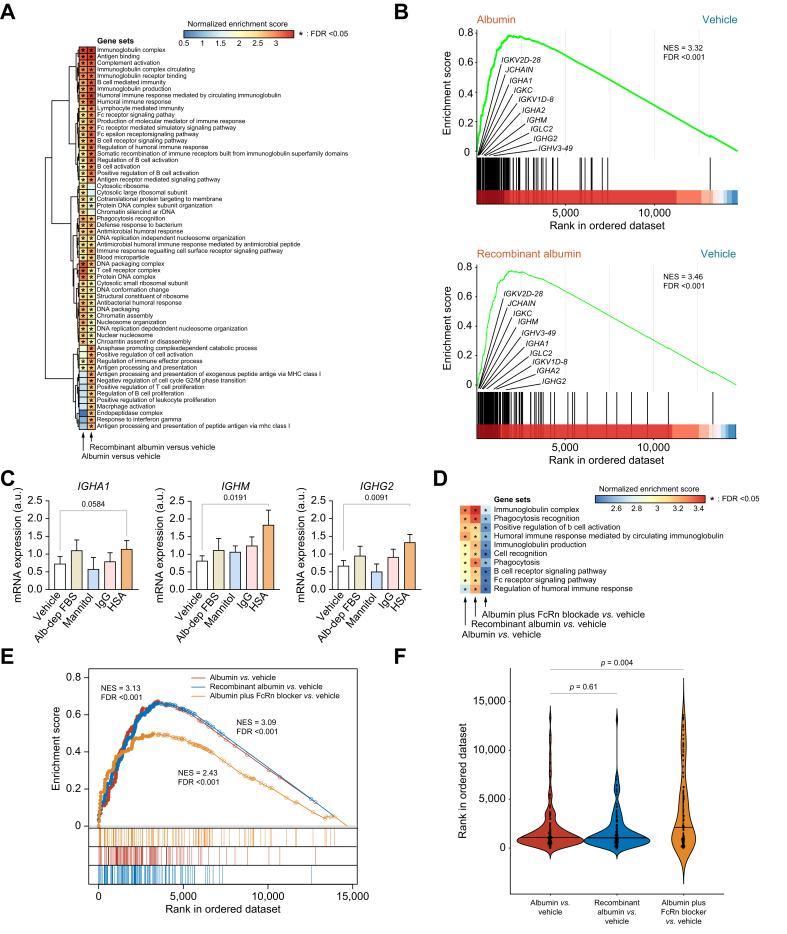
Fig. 2.
Changes in the transcriptional landscape of PBMCs isolated from patients with AD cirrhosis and incubated in vitro with HSA.
(A) GSEA was run on RNA-seq data to generate ranked lists of genes for the two following pairwise comparisons vs. vehicle: HSA and recombinant albumin (both at 15 mg/ml). Color gradient corresponds to increasing values of the NES of gene sets from the less (in blue) to the most (in red) upregulated. Asterisks in the heat map indicate FDR <0.05. (B) GSEA enrichment plots of the immunoglobulin complex gene set in two comparisons vs. vehicle: HSA (top) and recombinant albumin (bottom). The hash plot under GSEA curves shows where the members of the gene set appear in the ranked list of genes. The genes shown on the plots are representative of leading-edge genes (i.e. top-scoring genes). (C) Changes in the expression of three representative leading-edge genes coding for immunoglobulins in response to the vehicle control, 15% albumin-depleted FBS (Alb-dep FBS), mannitol (15 mg/ml), IgG (15 mg/ml), and HSA (15 mg/ml). Significant differences between groups were assessed using t tests. (D) Heat map of the NES for the 10 representative B cell‒related gene sets obtained for each of the following three comparisons vs. vehicle: HSA, recombinant albumin, and FcRn blocker (10 μg/ml) plus HSA vs. vehicle. (E) Enrichment plots of the immunoglobulin complex in the three GSEA comparisons described in D. The leading-edge genes are indicated by solid dots. The hash plots under GSEA curves show where the members of the gene set appear in each of the three ranked lists of genes. (F) Rank order of each gene of the immunoglobulin complex gene set. The higher in the rank, the lower the importance. Values of p are ranked from Kruskal–Wallis tests, followed by Mann–Whitney U tests. All FDR values are computed with adjustments for multiple testing and gene set size in the GSEA analysis. AD, acutely decompensated; FcRn, neonatal Fc receptor; FDR, false discovery rate; GSEA, gene set enrichment analysis; HSA, human serum albumin; HV, healthy volunteers; NES, normalized enrichment score; PBMC, peripheral blood mononuclear cell; RNA-seq, RNA sequencing.