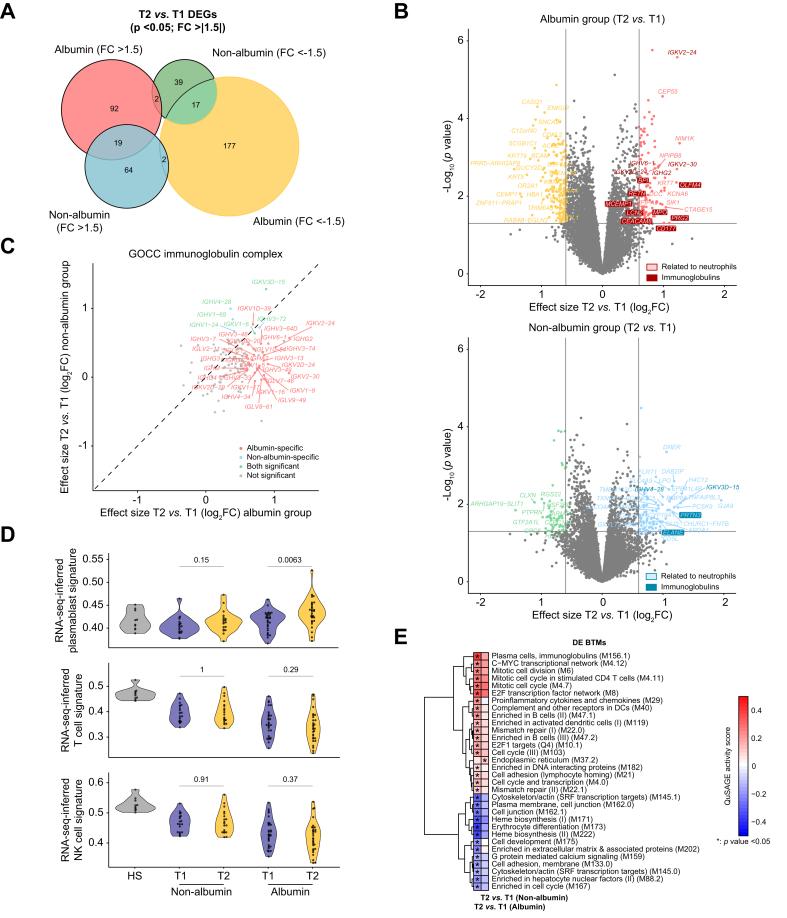
Fig. 5.
Whole blood gene signatures associated with HSA treatment.
(A) Euler plot showing DEGs between T2 and T1 for the albumin and non-albumin groups. DEGs were defined by an absolute FC greater than 1.5 and p <0.05. (B) Volcano plots showing differential expression effect size (log2 FC) between T2 and T1 plotted against significance (-log10p) for the albumin (top) and non-albumin (bottom) groups. In both volcano plots, gray points indicate genes with no significant difference in expression between T2 and T1 (with absolute FC <1.5 and p >0.05, i.e. -log10p <1.3). Colored points indicate DEGs, either upregulated or downregulated. The neutrophil-related genes are highlighted. (C) Differential expression effect size (log2 FC) between T2 and T1 in the albumin group compared with corresponding differential effect size in the non-albumin group. Only genes included in the Gene Ontology gene set ‘GOCC_Immunoglobulin Complex’ are shown here. DEGs with a concordant sign were considered as shared. (D) Violin plots of RNA-seq inferred signatures at T1 and T2 for plasmablasts, NK cells, and T cells, determined using the SingleR software, in the albumin and non-albumin groups. Baseline values in HV are also shown. Values of p are from Kruskal–Wallis tests. (E) Heat map of 32 DE BTMs between T2 and T1 that were specific for either the albumin or non-albumin group. Thirty-one DE BTMs were specific for the albumin group, whereas only one DE BTM (‘endoplasmic reticulum [M37.2]’) was specific for the non-albumin group. BTMs were hierarchically clustered based on QuSAGE activity scores obtained in the albumin group. Asterisks denote p <0.05 obtained when comparing probability density functions with QuSAGE. Color represents QuSAGE activity score. BTM, blood transcription module; DE, differentially expressed; DEG, differentially expressed gene; FC, fold change; HSA, human serum albumin; NK, natural killer; QuSAGE, Quantitative Set Analysis for Gene Expression; RNA-seq, RNA sequencing; T1, time 1; T2, time 2.