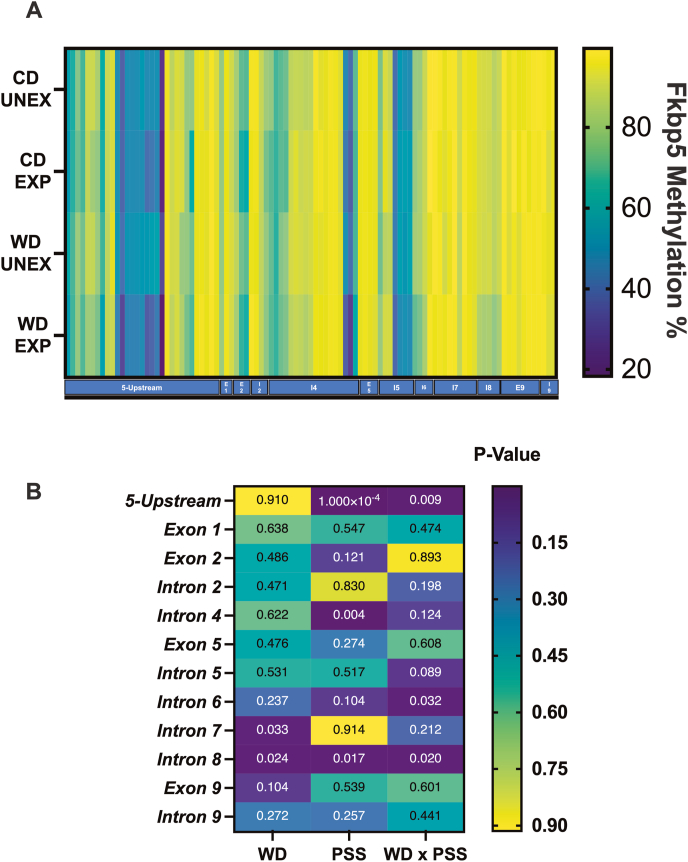
Fig. 8.
Heatmap and analysis of rat Fkbp5 methylation signatures per gene region. (A) Global percentages of Fkbp5 methylation per locus (B) Breakdown of significance per region. Methylation in the 5′-upstream region was significantly modified by PSS (p = 0.0001) and the interaction of WD x PPS (p = 0.009). Intron 4 was significantly modified by PSS (p = 0.003). Intron 6 was modified by WD x PSS (p = 0.032). Intron 7 was modified by the diet (p = 0.032); Intron 8 was modified by WD (p = 0.024), PSS (p = 0.017), and WD x PSS (0.024). The summary of statistical values of multiple comparisons in FKBP5 by regions can be found in Supplementary Table 16A–C and 17.