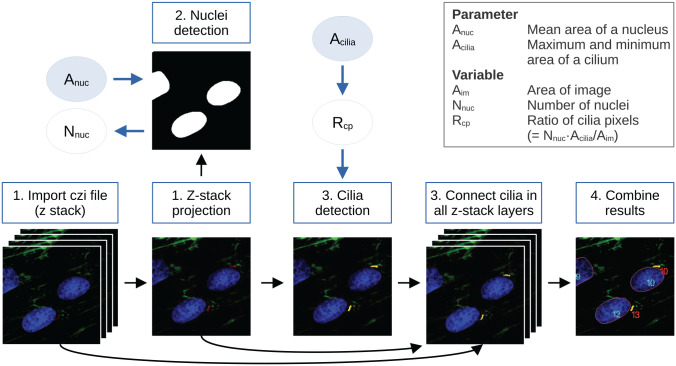
Figure 1.
Default workflow of detectCilia. 1. Image preparation: The original microscopy files are imported, and, if applicable, a maximum intensity z-stack projection is calculated. 2. Nuclei detection: The stained nuclei are segmented and counted using the mean area of a nucleus as an input parameter. 3. Cilia detection: Cilia are detected in the z-stack projection and every z-stack layer using the ratio of possible cilia pixels. Here, a primary cilium’s (expected) area is needed as an input parameter. 4. Cilia measurement: The information on all cilia (eg, lengths and heights) is collected, and the total lengths are calculated. The results are saved in a CSV file. (The images shown are sections of 190815_EV38_2_Collagen_ITSwithAsc+Dexa_63x_zstack_3.czi with enhanced brightness and contrast for better visualization.)