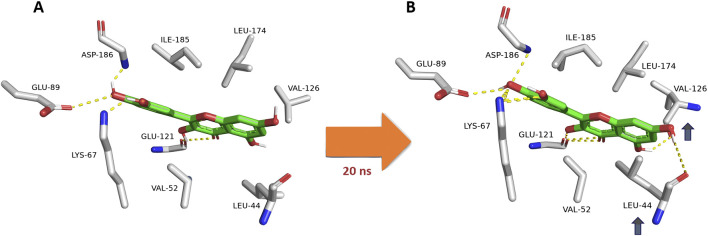
FIGURE 8.
Dynamic binding mode of quercetin (12) inside the Pim-1 kinase active site during its MD simulation. Quercetin (12) was chosen as a model for all derivatives containing hydroxyl group at C-3. (A) The initial binding mode of quercetin at 0 ns (Mode A binding pose). (B) After ≈20ns, Leu-44 and Val-126 became closer to C-5 and C-7 hydroxyl groups, respectively, where they established two stable H-bonds lasting to the end of 200-ns long MD simulation. In addition, both residues were able to sandwich the structure’s ring A via hydrophobic interactions. This observation did not occur with C-3 dehydroxylated derivatives, where its interactions with Glu-121 were transient.