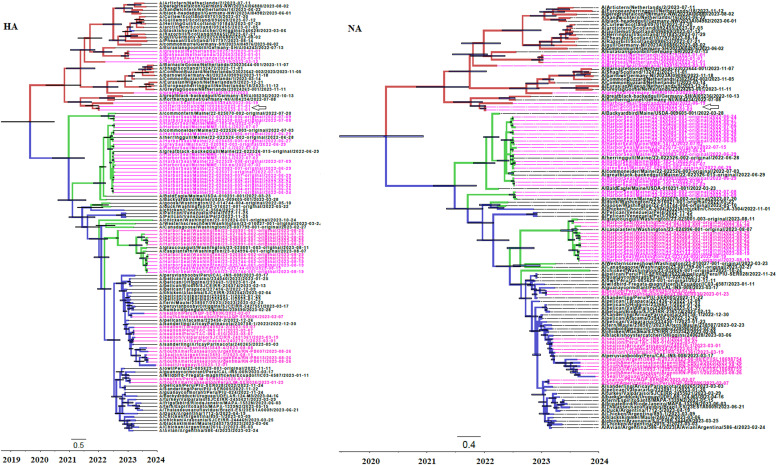
Fig. 1.
Bayesian MCMC phylogenetic tree analysis of HA and NA genes from HPIAV H5N1 2.3.4.4b strains. Maximum clade credibility trees are shown. Trees are rooted to the Most Recent Common Ancestor (MRCA). Time to the MRCA is shown in years at the bottom of the figure. Bar at the bottom of the trees denote time in years. Strains in the trees are shown by name followed by date of isolation. Clusters of strains isolated in Europe, North and South America are shown in red, green and blue, respectively. Node bars indicate the 95 % credibility values of the node heights. Strains isolated from birds are shown in black, while strains isolated from seals are shown in magenta. Strains isolated from otters are indicated by an arrow in magenta in both phylogenetic trees.