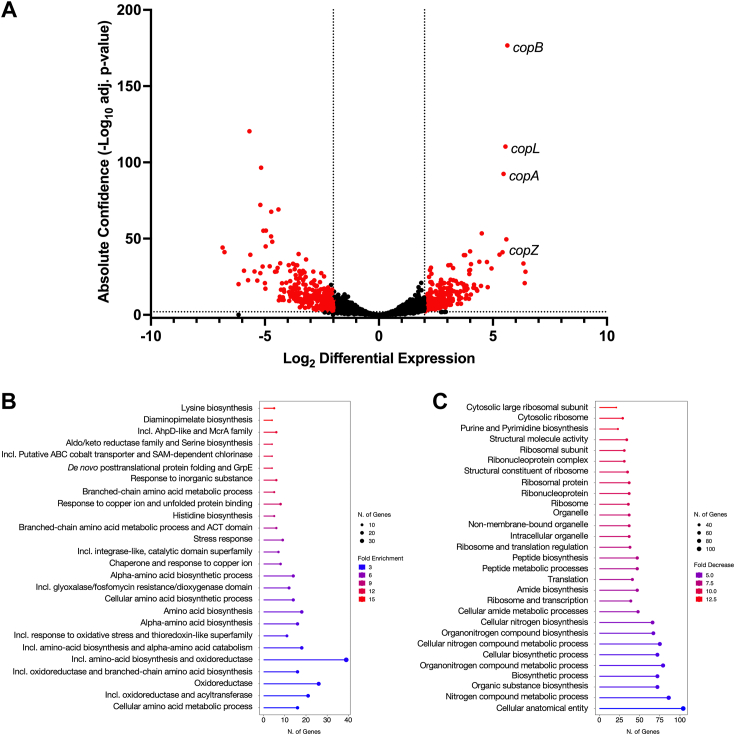
Figure 2.
R91 exposure results in the upregulation of copper homeostasis genes.A, volcano plot of RNAseq analysis of Staphylococcus aureus USA300 LAC in early exponential phase after 30 min exposure to sub-MIC R91 (3.125 μM). Results are from three independent experiments. Differentially expressed genes (absolute confidence >2, |log2 differential expression| >2) are colored in red. Detailed RNAseq data can be found in Table S3. Gene Ontology (GO) analysis of significantly upregulated (B) and downregulated (C) genes. Shown in (B and C) are the GO-enriched lollipop plots of differentially expressed pathways identified within the top 30 pathways. The GO terms corresponding to each pathway were initially filtered by a false discovery rate (FDR) of <0.05, before being selected by FDR and sorted for fold enrichment. The size of the dot represents the number of genes, and the color corresponds to the fold enrichment of the pathway.