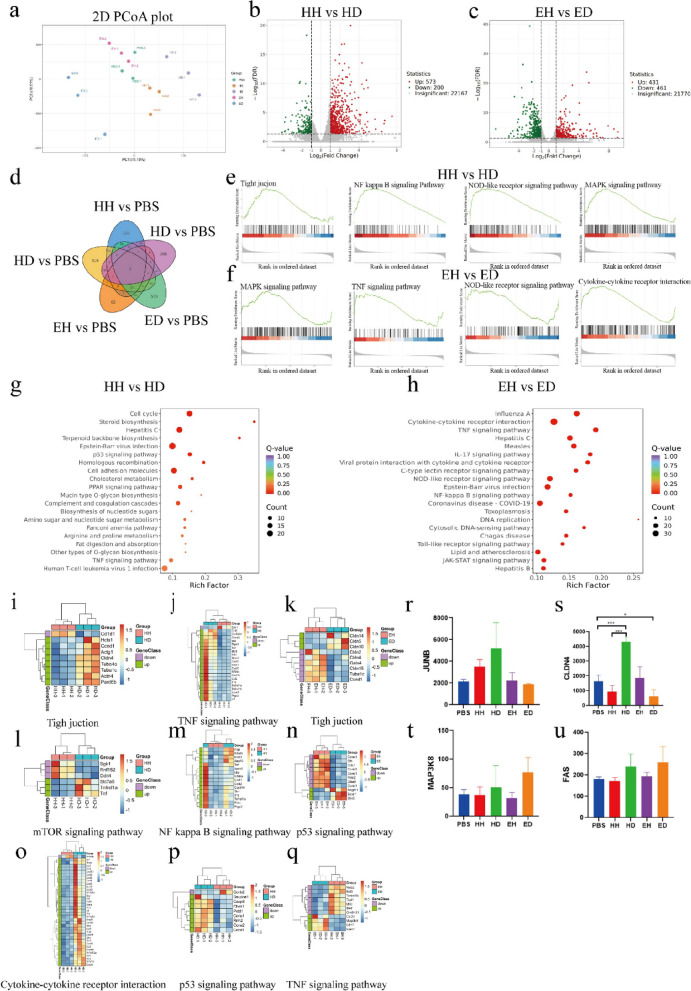
Fig. 5.
RNA-seq analysis was used to explore the mechanism underlying the disease resistance phenotype in mice. a PCA map of DEGs. b Volcano plot of HH and HD DEGs. c Volcano plot of EH and ED DEGs. d Venn diagram of DEGs. d GSEA diagram of HH and HD DEGs. f GSEA diagram of EH and ED DEGs. g KEGG enrichment map of HH and HD DEGs. h KEGG enrichment map of EH and ED DEGs. i Gene heatmap of the tight junction pathway in HH and HD. j Gene heatmap of the TNF signalling pathway in HH and HD. k Gene heatmap of the tight junction pathway in EH and ED. l Gene heatmap of the mTOR signalling pathway in HH and HD. m Gene heatmap of the NF-kappa B signalling pathway in HH and HD. n Gene heatmap of the p53 signalling pathway in EH and ED. o Gene heatmap of the cytokine‒cytokine receptor interaction in HH and HD. p Gene heatmap of the p53 signalling pathway in HH and HD. q Gene heatmap of the TNF signalling pathway in EH and ED. r Relative expression of JUNB. s The data are presented as the mean ± SEM (n = 6 mice per group). *P < 0.05, **P < 0.01, *** P < 0.001 were determined by one-way ANOVA with Bonferroni’s multiple comparisons test