Abstract
Background
Hepatocellular carcinoma (HCC) is a malignant tumor characterized by a high mortality rate. The occurrence and progression of HCC are linked to oxidative stress. Glyoxalase-1 (GLO1) plays an important role in regulating oxidative stress, yet the underlying mechanism remains unclear. GLO1 may serve as a prognostic biomarker and therapeutic target for HCC.
Methods
Based on TCGA database hepatocellular carcinoma samples, we conducted a bioinformatics analysis to explore the correlation between GLO1 expression and HCC cell proliferation and viability. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis revealed that differentially expressed genes (DEGs) were mainly enriched in the cell cycle pathway. We analyzed the relationships between GLO1 and 24 genes enriched in the cell cycle pathway using a protein-protein interaction (PPI) network. Finally, experimental validation was performed to assess GLO1’s impact on the distribution of cells at different cell cycle stages and on the proliferation and migration of HCC cells.
Results
Our study demonstrated that GLO1 was overexpressed in HCC tissues and was associated with a poor prognosis. Data analysis indicated that overexpression of GLO1 activated the cell cycle pathway and positively correlated with expression of the majority of key cell cycle genes. Experimental validation showed that GLO1 expression affects the number of HCC cells in G2 and S phases and regulates HCC cell proliferation and migration.
Conclusions
GLO1 represents a promising therapeutic target for HCC, providing valuable insights into its role in the viability and proliferation of HCC cells.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-024-12927-x.
Keywords: GLO1, Hepatocellular carcinoma, Cell cycle, Biomarker, Therapeutic target
Introduction
GLO1 is a cytoplasmic glutathione-dependent enzyme that detoxifies the glycolysis by-product methylglyoxal. Methylglyoxal is a cancer metabolite involved in metabolic reprogramming [1]. GLO1 plays a crucial role in tumorigenesis and the progression of various cancers. The mechanism of GLO1-mediated regulation of targeted cancer cells has garnered substantial attention [2–12]. While GLO1 is closely associated with cancer occurrence and development, its potential value as a therapeutic target for liver cancer treatment remains unexplored [13, 14]. Currently, comprehensive research on the role of GLO1 in HCC development is lacking, and existing findings require further validation.
According to global cancer statistics for 2020, liver cancer is the sixth-most common cancer in the world, with the third highest mortality rate and the fifth highest incidence [15]. Numerous factors contribute to liver cancer development, including hepatitis infections (HBV, HCV), alcohol consumption, obesity, diabetes mellitus, aflatoxin ingestion, non-alcoholic fatty liver disease, and metabolic syndrome [16–19]. Oxidative stress is a significant factor in carcinogenesis, playing a pivotal role in the development and progression of liver cancer of various etiologies [20–25]. Previous studies suggest that GLO1 is involved in the regulation of oxidative stress [26–29]. Given its role in oxidative stress regulation, GLO1 may be a potential target for cancer therapy [30].
This study aims to investigate the role of GLO1 in HCC and explore its potential as a prognostic biomarker and therapeutic target. We found that GLO1 expression levels were elevated in HCC tissues compared to normal tissues, with significant differences in survival associated with GLO1 expression. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis and Gene Ontology (GO) enrichment analysis were utilized to elucidate the biological processes and pathways associated with GLO1. Our results indicate that GLO1 contributes to the development of HCC by influencing the cell cycle pathway. Experimental verification further revealed that GLO1 predominantly affects the G2 and S phase of the HCC cell cycle and regulates HCC cell proliferation and migration. This study proposes a new therapeutic target for the treatment of HCC and clarifies the molecular mechanism of HCC development.
Methods
Data sources
GLO1 expression levels and clinical HCC data were obtained from The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov) [31]. Standardized TCGA and GTEX transcriptome data from normal tissue (N = 160) and tumor tissue (N = 374) samples were sourced from the UCSC database (https://xenabrowser.net/). The datasets GSE45436 (N = 39, T = 95), GSE57957 (N = 39, T = 39), and GSE76427 (N = 52, T = 115), containing liver cancer sequencing data, were downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/).
Analysis of GLO1 mutations in HCC
The cBioPortal online site is an open-source platform with multidimensional cancer genomic datasets and clinical data sources for exploration, visualization, and analysis [32]. We utilized the cBioPortal tool to analyze GLO1 mutations in HCC.
Survival analysis and HCC clinicopathological features
The “survival” and “survminer” R software packages were employed for survival analysis, and the Kaplan-Meier plotter assessed the association between GLO1 expression and patient survival in HCC [33]. Additionally, the clinicopathological data corresponding to the HCC samples were downloaded from TCGA and analyzed using R. The Kruskal-Wallis rank sum test identified significant differences (p < 0.05).
Relationship between GLO1 expression and immune cell infiltration
The ESTIMATE method inferred the ratio of stromal cells to immune cells in tumor samples, and the “estimate” R software package evaluated the stromal score, immune score, estimated score, and tumor purity [34]. The Tumor Immunity Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer/) enables systematic integrated correlation analysis of tumor-infiltrating immune cell features and selected pivotal genes [35]. We evaluated GLO1 expression levels in HCC tissues and analyzed their relationship with tumor purity and six types of infiltrating immune cells. Additionally, to explore the potential relationship between GLO1 expression and immune cell infiltration in HCC, we estimated the content of 22 immune cells using the CIBERSORT tool. Values of p < 0.05 were considered statistically significant.
Differentially expressed genes and pathway analysis
The HCC transcriptome data were analyzed using the limma package. The filter conditions to identify differentially expressed genes (DEGs) were as follows: fold change > |0.5|; adjusted p < 0.05. The Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) was used to perform GO and KEGG pathway enrichment analyses of DEGs.
Protein-protein interaction (PPI) network analysis
A protein-protein interaction (PPI) network graphically represents the physical and functional interactions between proteins in cells, constructed based on experimental data and computational predictions. NetworkAnalyst (https://www.networkanalyst.ca) is a tissue type–specific or cell type–specific protein-protein interaction (PPI) network analysis platform. The network integration algorithm uses the following bioinformatics tools: PPI networks, gene co-expression networks, and gene regulation networks [36]. We utilized NetworkAnalyst to construct a PPI network based on the relationships between GLO1 and proteins associated with the cell cycle.
Cell culture
Human Hep3B and Huh-7 cells were provided by the School of Biomedical Sciences at the Chinese University of Hong Kong. The cells were cultured in DMEM (Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific) and penicillin/streptomycin (1:100; Gibco, Thermo Fisher Scientific). All cells were grown at 37 °C in an incubator with 5% CO2 (SHEL LAB, USA).
Plasmid construction
GLO1-specific gRNA was designed using the Best CRISPR Design Tool for Knockouts (https://www.synthego.com/products/bioinformatics/crispr-design-tool) (TSINGKE, Chengdu, China). The process involved linearizing the plasmid backbone, annealing the gRNA, ligating the annealed oligonucleotides into the linearized vector, transforming the ligation product into Stbl3 cells, and verifying the gRNA sequence. The gRNA sequence is shown in Table 1. The overexpression plasmid was designed by Beijing Haichuangkeye Biological Technology Co., Ltd. (Beijing, China).
Table 1.
Primer sequences for GLOI-sgRNA
| gene | Primer sequences |
|---|---|
| GLO1-sgRNA-F | 5’-CACCGACTCTACTTCTTGGCTTATC-3’ |
| GLO1-sgRNA-R | 5’-AAACCATAAGCCAAGAAGTAGAGTC-3’ |
Construction of GLO1 knockout and overexpression cell models
The lentivirus packaging system, along with the helper plasmids PspAX2 and PMD2.G, was used to transfect the target plasmid into HEK293T cells. The virus-containing cell supernatant was collected 48 h and 72 h after transfection, mixed, and centrifuged to concentrate the virus particles using PEG8000. The knockout group and the negative control for the knockout group were denoted “sgGLO1” and “sgNC”, respectively; the overexpression group and its control were denoted “GLO1” and “control”, respectively. To obtain a stable cell line, HCC cells were screened using puromycin (2 µg/mL) 2 d after transfection.
Quantitative real-time polymerase chain reaction
Total RNA was extracted from HCC cells using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA). Reverse transcription was conducted using the FastKing RT Kit (TIANGEN, Chengdu, China) to produce cDNA. Quantitative real-time polymerase chain reaction (qRT-PCR; reaction volume: 10 µL) was performed using the 2× TSINGKE Master qPCR Mix (SYBR Green I) kit (TSINGKE, Beijing, China). 18s RNA was used as an internal reference. All primers were obtained from TSINGKE (Beijing, China; Table 2). mRNA expression levels were determined using the 2−∆∆Ct method.
Table 2.
The primer sequences
| Primer name | Forward Sequence (5’-3’) | Reverse Sequence (5’-3’) |
|---|---|---|
| GLO1 | CACTCTACTTCTTTGGCTTATGAGG | GGGTCTCATCATCTTCAGTGCC |
| 18sRNA | AAGTCCCTGCCCTTTGTACACA | GATCCGAGGGCCTCACTAAAC |
Western blot analysis
Total protein was extracted from HCC cells using 1 mM PMSF RIPA lysis buffer (Solarbio, China) and quantified using the BCA Protein Assay Kit (Beyotime, China). The protein lysate (80 µg) was denatured at 90 °C for 5 min in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) protein loading buffer (Beyotime, China). Samples were separated by 10% SDS-PAGE and transferred onto polyvinylidene fluoride membranes (Millipore, USA; IBFP0813C). The membranes were blocked with 5% non-fat milk in PBST for 1 h at room temperature and incubated overnight at 4 °C with primary anti-GLO1 antibody (1:1000 dilution, rabbit; Abcam, ab81461) and primary anti–β-actin antibody (1:2000 dilution, mouse; Beyotime, AF0003). The membranes were then washed three times with PBST and incubated with anti-rabbit IgG (1:2000 dilution; Beyotime; A0208) and anti-mouse IgG (1:2000 dilution; Beyotime; A0216) for 1 h at room temperature.
Cell proliferation assay
Cell suspensions from the four groups were inoculated into a 96-well culture plate at a density of 2000 cells per well and cultured at 37 °C in a 5% CO2 incubator. After 24, 48, 72, and 96 h of incubation, 90 µL of medium and 10 µL of cell counting kit-8 (CCK8) reagent (Dojindo, China) were added to each well. The cells were then incubated for an additional 2 h in the dark. The optical density of each well was measured at 460 nm using a microplate reader (Bio-Rad, Hercules, CA, USA, TY2018000102). Three wells were analyzed per group.
Wound healing assay
Cells (20 × 104) were seeded onto a six-well plate and incubated for 24 h until they reached confluence. The monolayer was scratched with a 10 µL pipette tip and washed with PBS to remove isolated cells. The cells were cultured in serum-free medium, and 4–5 images of the migration distance were captured in the same position for each well under an inverted fluorescence microscope (Nikon, Japan, Ts2R-FL) after 0, 24, and 48 h. The migration distance was calculated using ImageJ software.
Transwell invasion assay
The invasion assay was performed using a Transwell chamber (JETBIOFIL, Guangzhou, China) coated with Matrigel (Corning, Guangzhou, China). Transfected Hep3B and Huh-7 cells were suspended in serum-free medium and inoculated at a density of 2 × 104 cells per well into the upper chamber. The lower chamber contained medium supplemented with 10% FBS. After 48 h of incubation, the cells were fixed and stained with crystal violet. Photographs were taken under an inverted light microscope, and the invaded cells were counted using ImageJ software.
Flow cytometry analysis
Flow cytometry was performed to analyze the distribution of cells at different stages in the cell cycle. Cells in the logarithmic growth phase were inoculated uniformly into a six-well plate at a density of 2 × 104 cells per well and incubated in serum-free medium for 12 h. Next, the cells were transferred to complete medium and incubated for 48 h, after which they were washed with ice-cold PBS. Trypsin-digested cells were collected in a 1.5 mL conical tube and centrifuged at 1000 rpm for 3 min to obtain a precipitate. The cells were washed twice with ice-cold PBS and fixed with 75% ice-cold ethanol at -20℃ for 24 h. The cells were centrifuged, resuspended in PBS, treated with 50 µg/mL propidium iodide (PI) and 50 µg/mL RNase, and incubated at 37 °C for 30 min. Flow cytometry results (BD FACSVerseTM, USA) were analyzed using ModFit software.
In vivo experiments
Early zebrafish embryos (AB strain) were reared in Danien’s buffer containing 10 mmol/L phenylthiourea at 28.5 ℃ on a 10 h/14 h light/dark cycle. All HCC cell lines and culture conditions were established as previously described. HCC cells were stained with DiI (Biyuntian Biotech Co., Ltd, C1036) red fluorescent dye for 1 h before injection and then resuspended in PBS at a concentration of 1 × 104 cells/µL. Embryos were anesthetized by administering 0.05 mg/mL tricaine (ethyl 3-aminobenzoate methanesulfonate, Sigma). HCC cells (5–20 nL per embryo) were injected into the yolk space using a pneumatic pico pump syringe with a glass microinjection needle. Tumor size was quantified by fluorescence microscopy, and the cells were incubated for 3 days.
Statistical analysis
All experiments were repeated three times for accuracy. GraphPad Prism 7.0.0 software was used for paired group data analysis. Student’s t-test was used for statistical analysis. The threshold for statistical significance was set at p < 0.05.
Results
GLO1 protein is highly expressed in HCC
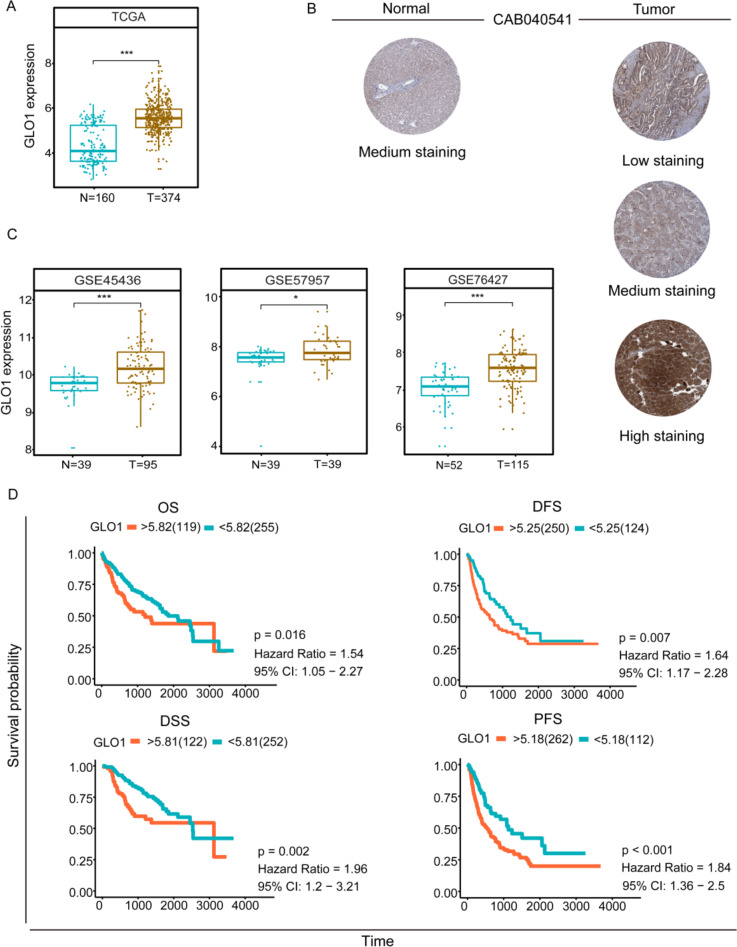
We analyzed GLO1 expression levels in HCC and normal tissues using mRNA expression data from TCGA and the GTEX database. We validated GLO1 expression in three GEO datasets. GLO1 mRNA expression was significantly higher in HCC samples than in normal tissues (Fig. 1A, C). Using the Human Protein Atlas Database, we compare GLO1 protein expression in HCC and normal tissues (Fig. 1B), confirming high GLO1 protein levels in HCC tissues.
Fig. 1.
GLO1 expression levels and survival analysis in HCC. (A) Comparison of GLO1 expression levels in HCC tissues and normal tissues was investigated by TCGA database. *p < 0.5, **p < 0.01, ***p < 0.001 between the two groups. (B) Protein levels from the Human Protein Atlas (HPA) assay GLO1 in normal and HCC tumor tissues. (C) GLO1 expression levels in HCC tissues compared to normal tissues verified by the GEO database. Between the two groups *p < 0.5, **p < 0.01, ***p < 0.001. (D) Kaplan-Meier survival analysis based on high and low GLO1 expression. overall survival (OS), disease-free survival (DFS), disease-specific survival (DSS), and progression-free survival (PFS)
GLO1 alterations in HCC
Based on DNA sequencing data, we identified the type and frequency of GLO1 mutations among HCC patients using the cBioPortal tool. The frequency of GLO1 alterations in HCC patients was 4%, including missense mutations (green), amplifications (red), and truncating mutations (light blue). Amplifications were the most frequent mutation type (Supplementary Fig. 2A). Supplementary Fig. 2B illustrates GLO1 mutation sites in HCC. Additionally, we investigated whether overall survival (OS) and disease-free survival (DFS) were associated with GLO1 mutations. HCC patients were divided into an altered group (n = 13) and an unaltered group (n = 353) based on cBioPortal data. Kaplan-Meier curves plotted OS and DFS (Supplementary Fig. 2C). GLO1 mutations were not significantly associated with OS (p = 0.266) or DFS (p = 0.722).
High GLO1 expression predicts poor prognosis in HCC patients
We aimed to verify if GLO1 expression could serve as a prognostic biomarker in HCC patients. We conducted OS, DFS, disease-specific survival (DSS), and progression-free survival (PFS) analyses of GLO1 in HCC using TCGA database. Kaplan-Meier analysis indicated that high GLO1 expression was significantly associated with poor prognosis (Fig. 1D). The relationship between GLO1 expression level and HCC stage, patient age, and patient sex was assessed using the Kruskal-Wallis rank sum test with Bonferroni adjustment. A significant association was found between GLO1 expression and stages 1 and 3 (adjusted p < 0.05), but no significant associations with other clinical factors (Supplementary Fig. 1). In conclusion, high GLO1 expression is a risk factor for poor prognosis in HCC patients.
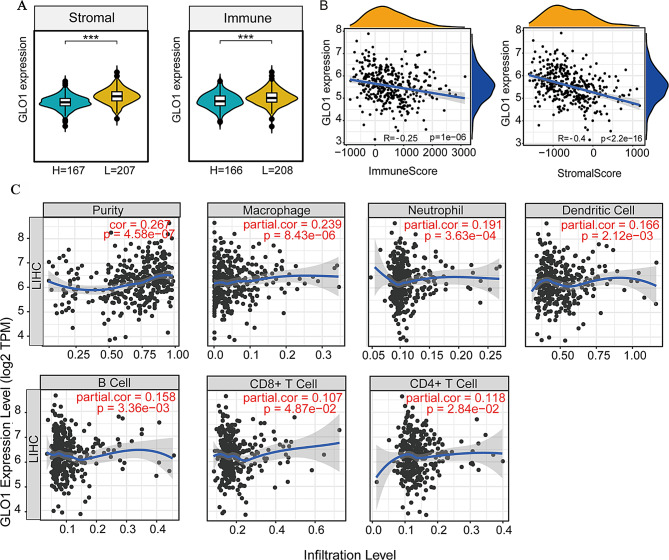
GLO1 is negatively and secondarily associated with immune infiltration in HCC
Oxidative stress can influence the immune response [37–40]. Given GLO1’s close association with oxidative stress, we hypothesized that GLO1 regulates the immune response and participates in immune surveillance. In addition, GLO1 participates in immune surveillance and immune cell infiltration control in another human malignancy [12]. GLO1 expression varied with HCC immune and stromal scores (Fig. 2A); it was negatively correlated with the immune score (r = -0.25, p = 1 × 10− 6) and stromal score (r = -0.4, p < 2.2 × 10− 6) (Fig. 2B). The correlation between GLO1 expression and stromal score was stronger than with the immune score. We then explored the relationship between GLO1 expression and immune cell infiltration levels. GLO1 expression showed a weak positive correlation with tumor purity (r = 0.267, p = 4.58 × 10− 7) and macrophage content (r = 0.239, p = 8.43 × 10− 6). GLO1 expression also exhibited a very weak positive correlation with the content of B cells (r = 0.158, p = 3.36 × 10− 3), CD8 + T cells (r = 0.107, p = 4.87 × 10− 2), CD4 + T cells (r = 0.118, p = 2.84 × 10− 2), neutrophils (r = 0.191, p = 3.63 × 10− 4) and dendritic cells (r = 0.166, p = 2.12 × 10− 3) (Fig. 2C). These results indicate that GLO1 plays a minor role in HCC immune cell infiltration.
Fig. 2.
Analysis of GLO1 expression and immune cell infiltration in HCC. (A) Association of GLO1 expression levels with differences in immune and stromal scores in HCC. Between the two groups *p < 0.5, **p < 0.01, ***p < 0.001. (B) Correlation analysis of GLO1 mRNA expression levels with immune score and stromal score in HCC. (C) The relationship between GLO1 gene expression and the level of infiltration of six types of immune cells in HCC was investigated using the Tumor Immune Estimation Resource (TIMER) database
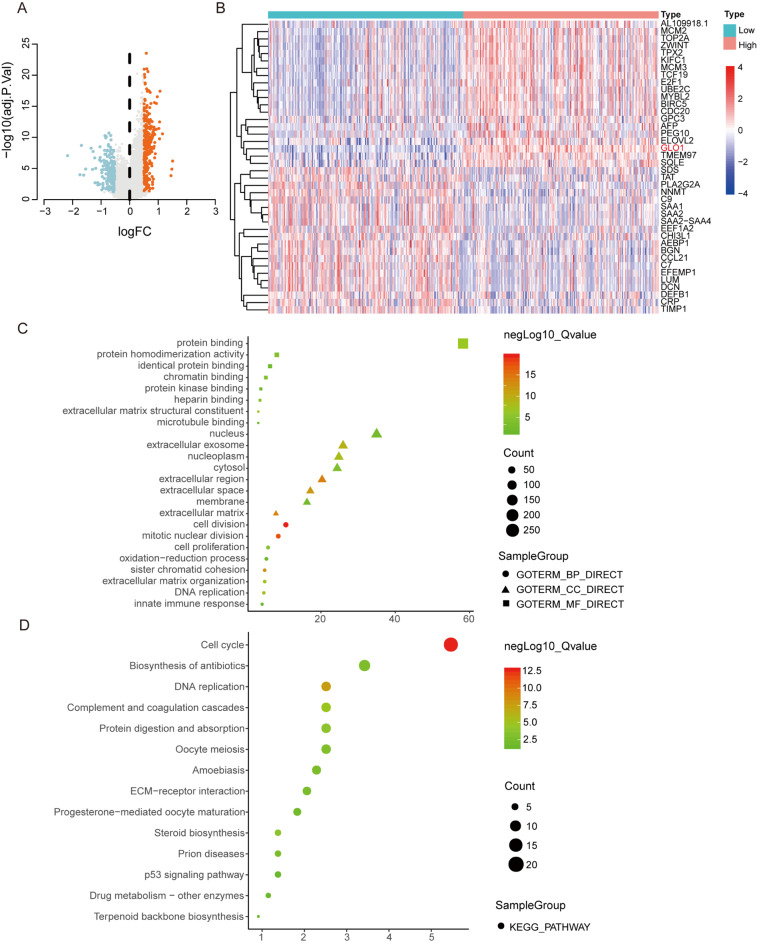
GLO1 expression is associated with tumor cell viability and proliferation
Patients were categorized into high and low groups based on GLO1 expression scores. Expression profiles were compared, revealing 458 DEGs related to GLO1. To elucidate the functional role of GLO1 in HCC, we performed functional enrichment analysis of 458 DEGs using the DAVID platform (Fig. 3A, B; Supplementary Table 1). GO enrichment analysis showed that, among molecular function (MF) terms, the DEGs were enriched in “protein binding” and “protein homodimerization activity”; among cellular component (CC) terms, they were enriched in “nucleus” and “extracellular exosome”; and among biological process (BP) terms, they were enriched in “cell division” and “mitotic nuclear division” (Fig. 3C and Supplementary Table2). KEGG pathway analysis revealed that the DEGs were mainly enriched in “cell cycle”, “biosynthesis of antibiotics”, and “DNA replication” (Fig. 3D and Supplementary Table 2).
Fig. 3.
Identification of DEGs in HCC and GO and KEGG enrichment analysis. (A) Volcano plots show the overall distribution of DEGs between the high and low GLO1 expression groups. (B) Heatmap of DEGs generated by comparison between the high and low GLO1 expression groups. (C) GO enrichment analysis of 458 DEGs was conducted via DAVID. GO terms are classified as biological process, cellular component, or molecular function terms. (D) KEGG pathway analysis revealed the signaling pathways in which the DEGs were enriched. Each point represents the enrichment level. The color corresponds to -log10 (adjusted p-value), and the size corresponds to the number of enriched genes
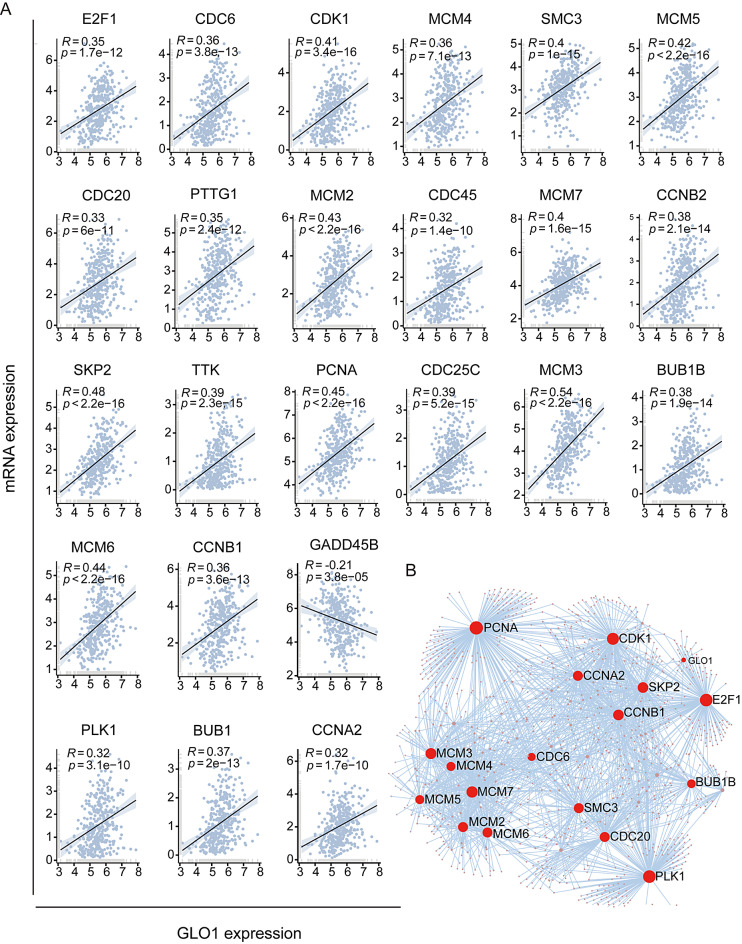
GLO1 regulates cell cycle pathway
To explore the transcription relationship between GLO1 and 24 genes enriched in the cell cycle pathway, we used the “ggpubr” package to perform correlation analysis at the transcriptional level in liver cancer. The expression of 23 genes was positively correlated with GLO1 expression: E2F1, CDC6, CDK1, SKP2, TTK, PCNA, CDC20, PTTG1, MCM2, CDC25C, MCM3, BUB1B, MCM4, SMC3, MCM5, MCM6, CCNB1, CDC45, MCM7, CCNB2, PLK1, BUBI, and CCNA2. GADD45B expression was negatively correlated with GLO1 expression (Fig. 4A). To further explore the potential interactions between these 24 genes and GLO1, a PPI network was constructed using the NetworkAnalyst tool (Fig. 4B).
Fig. 4.
Correlation analysis and PPI network analysis. (A) Correlation analysis of GLO1 and 24 genes enriched in the cell cycle pathway. (B) The PPI network of GLO1 and 24 cell cycle–related genes was constructed using the NetworkAnalysis tool
GLO1 knockout and overexpression affect the HCC cell cycle
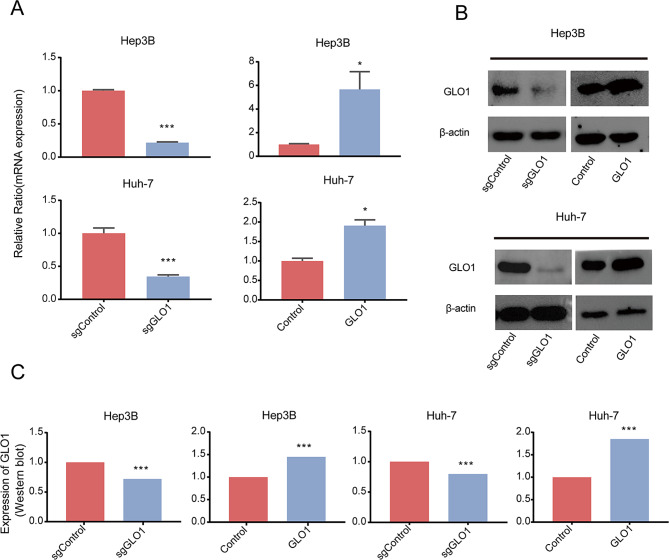
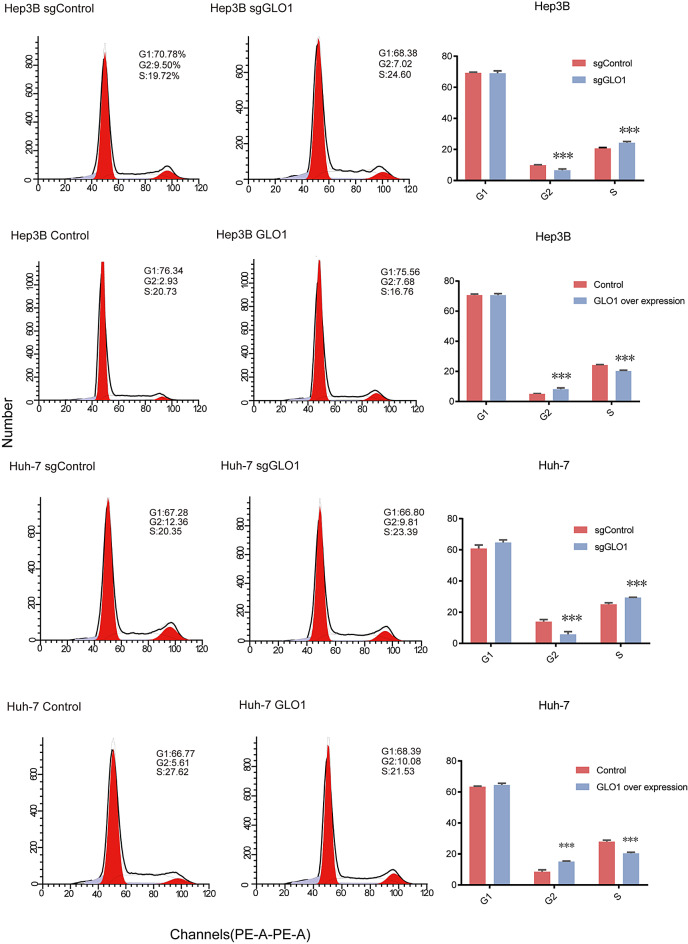
Bioinformatics analysis indicated a potential association between GLO1 expression and the cell cycle. To study the effect of GLO1 expression on the HCC cell cycle, we knocked out and overexpressed GLO1 in Hep3B and Huh-7 cells via lentiviral transduction and verified the efficiency of knockout and overexpression by qRT-PCR and western blotting (Fig. 5A, B, and C). We analyzed the distribution of cells in different cell cycle stages via flow cytometry and PI staining. Knockout and overexpression of GLO1 affected the number of Hep3B and Huh-7 cells in G2 and S phases. In GLO1-knockout cells, the number of cells in the G2 phase increased, the number of cells in the S phase decreased, and there was no significant change in the number of cells in the G1 phase. In GLO1-overexpressing cells, the number of cells in the G2 phase decreased, the number of cells in the S phase increased, and there was no significant change in the number of cells in the G1 phase (Fig. 6). These data indicated that GLO1 affected the number of HCC cells in G2 and S phases. Given that GLO1’s influence on these phases is critical for DNA replication and subsequent mitosis, we aim to further investigate whether GLO1 affects HCC cell proliferation and migration.
Fig. 5.
GLO1 knockout and overexpression in Hep3B and Huh-7 cells. (A) mRNA expression after GLO1 knockout and overexpression in Hep3B and Huh-7 cell lines was detected by qRT-PCR. (B, C) Protein expression after GLO1 knockout and overexpression in Hep3B and Huh-7 cell lines was determined by Western blot analysis (Original version in Supplementary Fig. 3). Statistical significance was determined by t-test: * p < 0.05; ** p < 0.01; *** p < 0.001
Fig. 6.
Effect of GLO1 knockout and overexpression of HCC cells in each cell cycle stage. GLO1 knockout and overexpression mainly affected the number of Hep3B and Huh-7 cells in G2 and S phases. Statistical significance was determined by t-test: * p < 0.05; ** p < 0.01; *** p < 0.001
GLO1 affects the proliferation, migration, and invasion of HCC cells by regulating the cell cycle
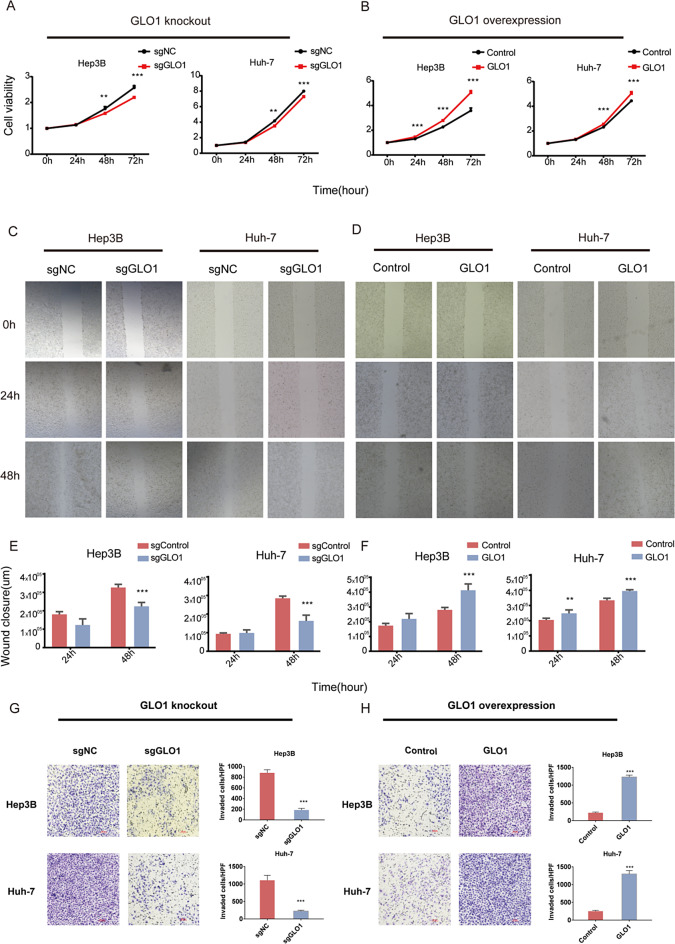
To determine how GLO1 affects cell proliferation and migration, we performed a CCK8 assay. GLO1 knockout significantly inhibited the proliferation of Hep3B and Huh-7 cells (Fig. 7A), whereas GLO1 overexpression enhanced proliferation (Fig. 7B). A wound healing assay was performed to analyze the effect of GLO1 on the migratory ability of HCC cells. We observed attenuated cell migration upon GLO1 knockout and enhanced cell migration upon GLO1 overexpression (Fig. 7C-F). According to the transwell invasion assay results, GLO1-overexpressing Hep3B and Huh-7 cells had significantly enhanced invasion ability, whereas GLO1-knockout cells exhibited significantly reduced invasion ability (Fig. 7G, H). Increased cell proliferation, migration and invasive capacity are key characteristic of cancer development and metastasis. Based on these in vitro findings, we proceeded to validate the role of GLO1 in HCC cells using an in vivo model.
Fig. 7.
GLO1 affects HCC cell proliferation, migration, and invasion by regulating the cell cycle pathway. (A, B) Proliferative capacity of GLO1-knockout and GLO1-overexpressing HCC cells was determined by CCK-8 assay. (C-F) Migration ability of GLO1-knockout and GLO1-overexpressing HCC cells was determined by wound healing assay. (G, H) Invasive ability of GLO1-knockout and GLO1-overexpressing HCC cells was determined by invasion assay. Statistical significance was determined by t-test: * p < 0.05; ** p < 0.01; *** p < 0.001
In vivo validation of the role of GLO1 in HCC cells
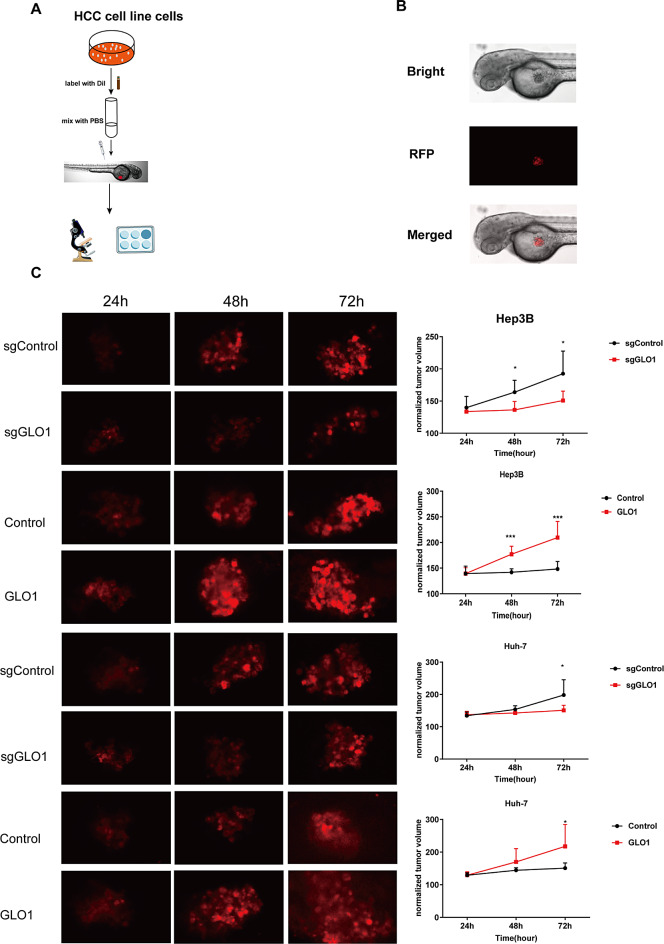
Transparent early zebrafish embryos (AB strain) were used to observe the changes in the growth of fluorescently labeled HCC cells in vivo (Fig. 8A). DiI labeled HCC cells injected into the yolk region of the zebrafish embryos were visualized using fluorescence microscopy (Fig. 8B). The proliferative ability of Hep3B and Huh-7 cells overexpressing GLO1 was significantly enhanced, whereas that of Hep3B and Huh-7 cells lacking GLO1 was significantly reduced (Fig. 8C). Based on these observations, GLO1 plays a critical role in promoting the proliferation of HCC cells in vivo, suggesting it as a potential therapeutic target for HCC.
Fig. 8.
GLO1 affects the growth of HCC cells in zebrafish. (A) Schematic depicting the procedure for constructing a zebrafish HCC xenograft model. (B) Fluorescence microscopy images of DiI stained HCC cells injected into the yolk region of zebrafish. Bright-field microscopy of whole zebrafish (AB line) 24 h after injection of human HCC cells (upper panel). Fluorescence microscopy of DiI stained HCC cells under red fluorescent protein (RFP) channel (middle). Merged bright-field and RFP field microscopy images (bottom). (C) Growth of DiI stained HCC cell xenografts in zebrafish
Discussion
In recent years, the incidence of HCC has risen significantly, making it one of the most common cancers. High GLO1 expression plays a crucial role in tumor initiation and progression [3, 5, 6, 8, 9, 13, 41]. GLO1 is upregulated in HCC and plays an essential role in HCC cell proliferation, thus presenting itself as a potential therapeutic target [13, 14]. However, comprehensive research on the association between GLO1 expression and the occurrence and development of HCC remains sparse. This study comprehensively investigated the role of GLO1 in hepatocellular carcinoma (HCC) and its potential as a therapeutic target. This study investigates the role of GLO1 in HCC and its potential as a therapeutic target through bioinformatics analysis of public data.
Using TCGA data, we evaluated GLO1 expression in HCC tissues and confirmed that GLO1 transcription levels were significantly elevated in tumor tissues compared to adjacent normal tissues (Fig. 1A, B, and C), aligning with previous findings [13, 14]. We further examined GLO1 mutations in HCC at the DNA level, revealing a mutation rate of 4%, though these mutations were not significantly associated with survival (Supplementary Fig. 2). To assess the impact of GLO1 expression on survival, we analyzed four types of survival data: OS, DFS, DSS, and PFS. GLO1 expression was significantly associated with the survival rate of HCC patients, suggesting its potential as a prognostic indicator. GLO1’s role in promoting tumor proliferation and survival has been highlighted in multiple cancer types, including prostate cancer, breast cancer, pancreatic cancer, malignant melanoma, and gastric cancer [3, 6, 9–12, 41, 42]. GLO1 expression was not significantly associated with clinicopathological parameters such as grade, age, and sex (Supplementary Fig. 1). GLO1 is linked to oxidative stress [26], which affects the immune response [30, 37–40]. We speculated that GLO1 also regulates tumor immunity and found that GLO1 plays a minor role in HCC immune cell infiltration (Fig. 2).
To investigate the molecular functions of GLO1 in HCC, we performed GO and KEGG pathway enrichment analysis of DEGs. Enriched BP terms included “cell division” and “mitotic nuclear division” (Fig. 3C). KEGG pathway analysis revealed that the DEGs were mainly enriched in “cell cycle” and “DNA replication” (Fig. 3D). GLO1 promotes tumor proliferation [3, 10, 14], though the exact mechanism remains unclear. We conducted a correlation analysis between GLO1 expression and genes related to the cell cycle pathway. Specifically, the expression of E2F1, CDC6, CDK1, SKP2, TTK, PCNA, CDC20, PTTG1, MCM2, CDC25C, MCM3, BUB1B, MCM4, SMC3, MCM5, MCM6, CCNB1, CDC45, MCM7, CCNB2, PLK1, BUBI, and CCNA2 was positively correlated with GLO1 expression, whereas GADD45B expression was negatively correlated (Fig. 4A). Given that alterations in cell cycle regulation are known to drive tumor proliferation and survival by enabling cancer cells to bypass normal growth constraints and evade apoptosis [43]. We hypothesize that GLO1 may contribute to tumor proliferation and viability by activating the cell cycle pathway. GLO1 expression was significantly correlated with that of genes enriched in the cell cycle pathway (Fig. 4A). However, this does not establish a causal relationship between GLO1 and cell cycle pathway activation. Therefore, we constructed a GLO1 knockout and overexpression system for further validation. GLO1 expression predominantly influenced the number of cells in G2 and S phase of the cell cycle (Fig. 6), supporting previous reports that GLO1’s role in tumor proliferation and survival [5, 7, 8, 14]. The results demonstrated that GLO1 affects the proliferation and migration of HCC cells by regulating the cell cycle (Fig. 7).
In conclusion, this study provides comprehensive evidence supporting GLO1’s pivotal role in HCC development. Our findings indicate that GLO1 is overexpressed in HCC tissues and is associated with a poor prognosis, similar to other human malignancies [44]. Furthermore, our findings elucidate that overexpression of GLO1 activates the cell cycle pathway and correlates with genes enriched in this pathway. Experimental validation indicated that GLO1 expression affects the number of HCC cells in G2 and S phases and regulates HCC cell proliferation and migration. These results underscore GLO1 as a promising therapeutic target for HCC, offering valuable insights into its role in tumor viability and proliferation. Future directions include further elucidation of the underlying molecular mechanisms and validation of GLO1-targeted therapeutic strategies in clinical settings.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Abbreviations
- DFS
Disease-free survival
- DSS
Disease-specific survival
- GEO
Gene Expression Omnibus
- GO
Gene Ontology
- GLO1
Glyoxalase-1
- HBV
Hepatitis B virus
- HCC
Hepatocellular carcinoma
- HCV
Hepatitis C virus
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- OS
Overall survival
- PFS
Progression-free survival
- PPI
Protein-protein interaction
- TCGA
The Cancer Genome Atlas
Author contributions
F.K.D, Z.G.X, and L.Y, Designed the experiments. Y.Z, X.L.T, L.L, D.C, S.G, S.Y.H, and Y.L, Analyzed the data and performed the experiments. J.S, Y.C, Y.S.Z, X.W, M.X.L, M.J.C, X.B.L, Y.H.S, L.G, W.P.L, F.W, Z.Z, X.D.W, and S.D, Supervised the study and coordinated the writing of the manuscript.
Funding
This study was supported by grants from the Sichuan Science and Technology Program, China (No. 2022NSFSC0783, No. 2022YFS0619), the Joint Founds of Southwest Medical University and Luzhou Government (No.2020LZXNYDJ08), Grants of Southwest Medical University (2021ZKMS038), Funds of talent introduction and scientific research of Southwest Medical University (No.05-00040140), The Project of Science and Technology Department of Sichuan Provincial of China to L.Y. (2019JDJQ0035).
Data availability
GLO1 expression levels and clinical HCC data were obtained from The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov) [31]. Standardized TCGA and GTEX transcriptome data from normal tissue (N = 160) and tumor tissue (N = 374) samples were retrieved from the UCSC database (https://xenabrowser.net/). The datasets GSE45436 (N = 39, T = 95), GSE57957 (N = 39, T = 39), and GSE76427 (N = 52, T = 115), containing liver cancer sequencing data, were downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/).
Declarations
Ethics approval and consent to participate
The study was performed in accordance with the relevant guidelines and regulations. All animal protocols were approved by the Ethics Committee of Southwest Medical University (Date: February 24, 2021/No.: 20210223-243). This research was in accordance with ARRIVE guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yao Zhang, Xiaolong Tang and Lin Liu contributed equally to this work and share first authorship.
Contributor Information
Zhangang Xiao, Email: zhangangxiao@swmu.edu.cn.
Lei Yao, Email: yaolei2009@gmail.com.
Fukuan Du, Email: adublg@126.com.
References
- 1.Jandova J, Wondrak GT. Genomic GLO1 deletion modulates TXNIP expression, glucose metabolism, and redox homeostasis while accelerating human A375 malignant melanoma tumor growth. Redox Biol. 2021;39:101838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiavarina B, Nokin MJ, Bellier J, Durieux F, Bletard N, Sherer F, Lovinfosse P, Peulen O, Verset L, Dehon R et al. Methylglyoxal-mediated stress correlates with high metabolic activity and promotes Tumor Growth in Colorectal Cancer. Int J Mol Sci 2017;18(1). [DOI] [PMC free article] [PubMed]
- 3.Hutschenreuther A, Bigl M, Hemdan NY, Debebe T, Gaunitz F, Birkenmeier G. Modulation of GLO1 expression affects malignant properties of cells. Int J Mol Sci 2016;17(12). [DOI] [PMC free article] [PubMed]
- 4.Luengo A, Abbott KL, Davidson SM, Hosios AM, Faubert B, Chan SH, Freinkman E, Zacharias LG, Mathews TP, Clish CB, et al. Reactive metabolite production is a targetable liability of glycolytic metabolism in lung cancer. Nat Commun. 2019;10(1):5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian X, Wang Y, Ding X, Cheng W. High expression of GLO1 indicates unfavorable clinical outcomes in glioma patients. J Neurosurg Sci 2019. [DOI] [PubMed]
- 6.Burdelski C, Shihada R, Hinsch A, Angerer A, Gobel C, Friedrich E, Hube-Magg C, Burdak-Rothkamm S, Kluth M, Simon R, et al. High-level glyoxalase 1 (GLO1) expression is linked to poor prognosis in prostate cancer. Prostate. 2017;77(15):1528–38. [DOI] [PubMed] [Google Scholar]
- 7.Santarius T, Bignell GR, Greenman CD, Widaa S, Chen L, Mahoney CL, Butler A, Edkins S, Waris S, Thornalley PJ, et al. GLO1-A novel amplified gene in human cancer. Genes Chromosomes Cancer. 2010;49(8):711–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bair WB 3rd, Cabello CM, Uchida K, Bause AS, Wondrak GT. GLO1 overexpression in human malignant melanoma. Melanoma Res. 2010;20(2):85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Kuramitsu Y, Ueno T, Suzuki N, Yoshino S, Iizuka N, Akada J, Kitagawa T, Oka M, Nakamura K. Glyoxalase I (GLO1) is up-regulated in pancreatic cancerous tissues compared with related non-cancerous tissues. Anticancer Res. 2012;32(8):3219–22. [PubMed] [Google Scholar]
- 10.Cheng WL, Tsai MM, Tsai CY, Huang YH, Chen CY, Chi HC, Tseng YH, Chao IW, Lin WC, Wu SM, et al. Glyoxalase-I is a novel prognosis factor associated with gastric cancer progression. PLoS ONE. 2012;7(3):e34352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antognelli C, Marinucci L, Frosini R, Macchioni L, Talesa VN. Metastatic prostate Cancer cells secrete methylglyoxal-derived MG-H1 to reprogram human osteoblasts into a dedifferentiated, malignant-like phenotype: a possible novel player in prostate Cancer bone metastases. Int J Mol Sci 2021, 22(19). [DOI] [PMC free article] [PubMed]
- 12.Antognelli C, Mandarano M, Prosperi E, Sidoni A, Talesa VN. Glyoxalase-1-Dependent methylglyoxal depletion sustains PD-L1 expression in metastatic prostate Cancer cells: a novel mechanism in Cancer Immunosurveillance escape and a potential Novel Target to Overcome PD-L1 Blockade Resistance. Cancers 2021, 13(12). [DOI] [PMC free article] [PubMed]
- 13.Zhang S, Liang X, Zheng X, Huang H, Chen X, Wu K, Wang B, Ma S. Glo1 genetic amplification as a potential therapeutic target in hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7(5):2079–90. [PMC free article] [PubMed] [Google Scholar]
- 14.Hu X, Yang X, He Q, Chen Q, Yu L. Glyoxalase 1 is up-regulated in hepatocellular carcinoma and is essential for HCC cell proliferation. Biotechnol Lett. 2014;36(2):257–63. [DOI] [PubMed] [Google Scholar]
- 15.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. [DOI] [PubMed] [Google Scholar]
- 16.Donato F, Gelatti U, Tagger A, Favret M, Ribero ML, Callea F, Martelli C, Savio A, Trevisi P, Nardi G. Intrahepatic cholangiocarcinoma and hepatitis C and B virus infection, alcohol intake, and hepatolithiasis: a case-control study in Italy. Cancer Causes Control: CCC. 2001;12(10):959–64. [DOI] [PubMed] [Google Scholar]
- 17.Marengo A, Rosso C, Bugianesi E. Liver Cancer: connections with obesity, fatty liver, and cirrhosis. Annu Rev Med. 2016;67:103–17. [DOI] [PubMed] [Google Scholar]
- 18.Grandhi MS, Kim AK, Ronnekleiv-Kelly SM, Kamel IR, Ghasebeh MA, Pawlik TM. Hepatocellular carcinoma: from diagnosis to treatment. Surg Oncol. 2016;25(2):74–85. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Wang H. Heterogeneity of liver cancer and personalized therapy. Cancer Lett. 2016;379(2):191–7. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Li Z, Ye Y, Xie L, Li W. Oxidative Stress and Liver Cancer: Etiology and Therapeutic Targets. Oxid Med Cell Longev. 2016;2016:7891574. [DOI] [PMC free article] [PubMed]
- 21.Shen J, Chen M, Lee D, Law CT, Wei L, Tsang FH, Chin DW, Cheng CL, Lee JM, Ng IO, et al. Histone chaperone FACT complex mediates oxidative stress response to promote liver cancer progression. Gut. 2020;69(2):329–42. [DOI] [PubMed] [Google Scholar]
- 22.Liu MX, Jin L, Sun SJ, Liu P, Feng X, Cheng ZL, Liu WR, Guan KL, Shi YH, Yuan HX, et al. Metabolic reprogramming by PCK1 promotes TCA cataplerosis, oxidative stress and apoptosis in liver cancer cells and suppresses hepatocellular carcinoma. Oncogene. 2018;37(12):1637–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLoughlin MR, Orlicky DJ, Prigge JR, Krishna P, Talago EA, Cavigli IR, Eriksson S, Miller CG, Kundert JA, Sayin VI, et al. TrxR1, gsr, and oxidative stress determine hepatocellular carcinoma malignancy. Proc Natl Acad Sci USA. 2019;116(23):11408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee D, Xu IM, Chiu DK, Leibold J, Tse AP, Bao MH, Yuen VW, Chan CY, Lai RK, Chin DW, et al. Induction of oxidative stress through inhibition of Thioredoxin Reductase 1 is an effective Therapeutic Approach for Hepatocellular Carcinoma. Hepatology. 2019;69(4):1768–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marra M, Sordelli IM, Lombardi A, Lamberti M, Tarantino L, Giudice A, Stiuso P, Abbruzzese A, Sperlongano R, Accardo M, et al. Molecular targets and oxidative stress biomarkers in hepatocellular carcinoma: an overview. J Translational Med. 2011;9:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Zhang J, Chen L, Li J, Zhang H, Guo X. Glycine Suppresses AGE/RAGE Signaling Pathway and Subsequent Oxidative Stress by Restoring Glo1 Function in the Aorta of Diabetic Rats and in HUVECs. Oxid Med Cell Longev. 2019;2019:4628962. [DOI] [PMC free article] [PubMed]
- 27.Antognelli C, Trapani E, Delle Monache S, Perrelli A, Fornelli C, Retta F, Cassoni P, Talesa VN, Retta SF. Data in support of sustained upregulation of adaptive redox homeostasis mechanisms caused by KRIT1 loss-of-function. Data Brief. 2018;16:929–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delle Monache S, Pulcini F, Frosini R, Mattei V, Talesa VN, Antognelli C. Methylglyoxal-dependent glycative stress is prevented by the natural antioxidant oleuropein in Human Dental Pulp Stem Cells through Nrf2/Glo1 pathway. Antioxidants 2021, 10(5). [DOI] [PMC free article] [PubMed]
- 29.Gambelunghe A, Giovagnoli S, Di Michele A, Boncompagni S, Dell’Omo M, Leopold K, Iavicoli I, Talesa VN, Antognelli C. Redox-Sensitive glyoxalase 1 Up-Regulation is crucial for protecting human lung cells from gold nanoparticles toxicity. Antioxidants 2020;9(8). [DOI] [PMC free article] [PubMed]
- 30.Du F, Li Y, Shen J, Zhao Y, Kaboli PJ, Xiang S, Wu X, Li M, Zhou J, Zheng Y, et al. Glyoxalase 1 gene improves the antistress capacity and reduces the immune inflammatory response. BMC Genet. 2019;20(1):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol. 2015;19(1A):A68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu P, Heins ZJ, Muller JT, Katsnelson L, de Bruijn I, Abeshouse AA, Schultz N, Fenyo D, Gao J. Integration and analysis of CPTAC Proteomics Data in the context of Cancer Genomics in the cBioPortal. Mol Cell Proteomics: MCP. 2019;18(9):1893–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Therneau T, Grambsch P. Modeling Survival Data: extending the Cox Model. New York, NY: Springer; 2000. [Google Scholar]
- 34.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Trevino V, Shen H, Laird PW, Levine DA, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: a web server for Comprehensive Analysis of Tumor-infiltrating Immune cells. Cancer Res. 2017;77(21):e108–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou G, Soufan O, Ewald J, Hancock REW, Basu N, Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47(W1):W234–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Datta S, Cano M, Ebrahimi K, Wang L, Handa JT. The impact of oxidative stress and inflammation on RPE degeneration in non-neovascular AMD. Prog Retin Eye Res. 2017;60:201–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dietz RM, Wright CJ. Oxidative stress diseases unique to the perinatal period: a window into the developing innate immune response. Am J Reprod Immunol. 2018;79(5):e12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laddha NC, Dwivedi M, Mansuri MS, Gani AR, Ansarullah M, Ramachandran AV, Dalai S, Begum R. Vitiligo: interplay between oxidative stress and immune system. Exp Dermatol. 2013;22(4):245–50. [DOI] [PubMed] [Google Scholar]
- 40.Zhang B, Zehnder JL. Oxidative stress and immune thrombocytopenia. Semin Hematol. 2013;50(3):e1–4. [DOI] [PubMed] [Google Scholar]
- 41.Lv N, Hao S, Luo C, Abukiwan A, Hao Y, Gai F, Huang W, Huang L, Xiao X, Eichmuller SB, et al. miR-137 inhibits melanoma cell proliferation through downregulation of GLO1. Sci China Life Sci. 2018;61(5):541–9. [DOI] [PubMed] [Google Scholar]
- 42.Antognelli C, Cecchetti R, Riuzzi F, Peirce MJ, Talesa VN. Glyoxalase 1 sustains the metastatic phenotype of prostate cancer cells via EMT control. J Cell Mol Med. 2018;22(5):2865–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411(6835):342–8. [DOI] [PubMed] [Google Scholar]
- 44.Antognelli C, Talesa VN. Glyoxalases in Urological malignancies. Int J Mol Sci 2018, 19(2). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GLO1 expression levels and clinical HCC data were obtained from The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov) [31]. Standardized TCGA and GTEX transcriptome data from normal tissue (N = 160) and tumor tissue (N = 374) samples were retrieved from the UCSC database (https://xenabrowser.net/). The datasets GSE45436 (N = 39, T = 95), GSE57957 (N = 39, T = 39), and GSE76427 (N = 52, T = 115), containing liver cancer sequencing data, were downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/).