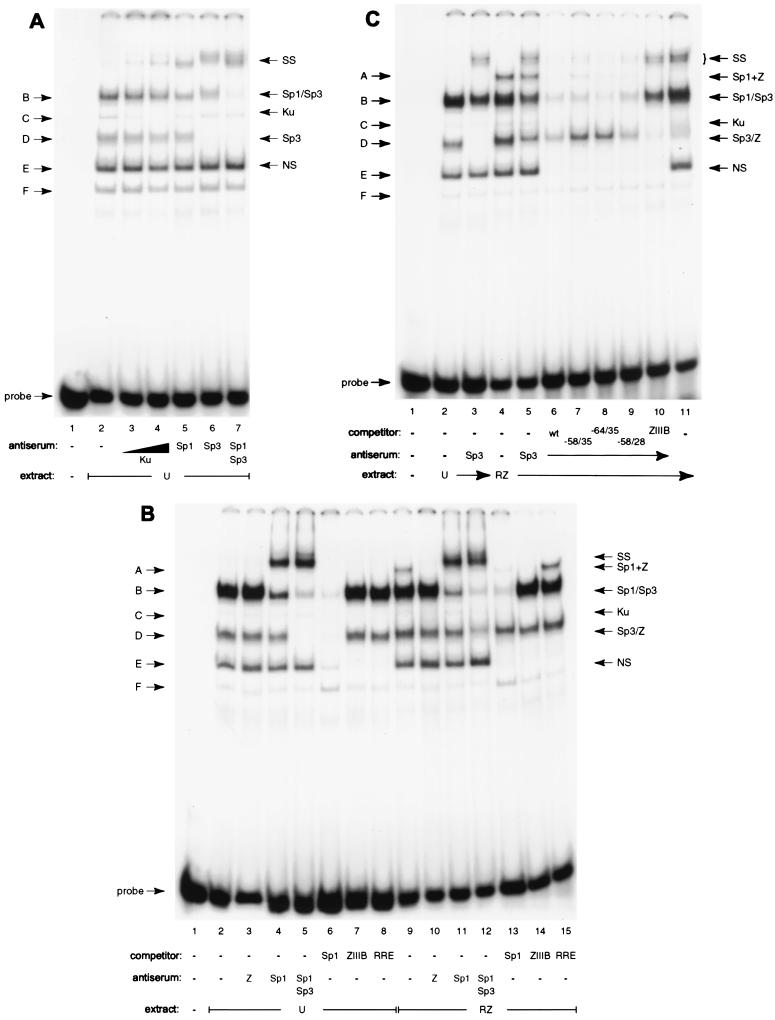
FIG. 7.
Identification of proteins binding to Rp −64/−28 by antibody supershifts and by oligonucleotide competition. (A) Identification of proteins in uninduced (U) Cl16 extracts binding to the Rp oligonucleotide probe. Complex designation is indicated on the left (B to F); identified proteins are on the right (SS, supershifted bands). Lane 1, probe alone; lanes 2 to 7, 7 μg of cell extract; lanes 3 to 7 contain antiserum to the indicated proteins. (B) Sp1 and ZEBRA occupy Rp −64/−28 together in complex A. Extracts of uninduced cells (U) and of Cl16 cells transfected with ZEBRA and Rta expression vectors (RZ) were incubated with the oligonucleotide probe and the indicated antisera (lanes 3 to 5 and 10 to 12) or a 500-fold excess of the indicated unlabeled competitor DNA (lanes 6 to 8 and 14 to 16). (C) Competition and supershift analysis identify ZEBRA in complex D1 in extracts of induced Cl16 cells. Uninduced (U, lanes 2 and 3) and Rta and ZEBRA-transfected (RZ, lanes 4 to 11) Cl16 extracts were incubated with the Rp probe and Sp3 (lanes 3 and 5 to 10) and ZEBRA (lane 11) antisera. Binding reactions in lanes 6 to 10 also contained a 500-fold excess of the indicated unlabeled competitor DNA (lanes 6 to 9 contained Rp −64/−28, −58/−28, −64/−35, and −58/−35, respectively; lane 10, ZRE ZIIIB).