Abstract
Introduction
Efforts to harmonize lipidomic methodologies have been limited within the community. Here, we aimed to capitalize on the recent National Institute of Standards and Technology lipidomics interlaboratory comparison exercise by implementing a questionnaire that assessed current methodologies, quantitation strategies, standard operating procedures (SOPs), and quality control activities employed by the lipidomics community.
Objectives
Lipidomics is a rapidly developing field with diverse applications. At present, there are no community-vetted methods to assess measurement comparability or data quality. Thus, a major impetus of this questionnaire was to profile current efforts, highlight areas of need, and establish future objectives in an effort to harmonize lipidomics workflows.
Methods
The 54-question survey inquired about laboratory demographics, lipidomic methodologies and SOPs, analytical platforms, quantitation, reference materials, quality control procedures, and opinions regarding challenges existing within the community.
Results
A total of 125 laboratories participated in the questionnaire. A broad overview of results highlighted a wide methodological diversity within current lipidomic workflows. The impact of this diversity on lipid measurement and quantitation is currently unknown and needs to be explored further. While some laboratories do incorporate SOPs and quality control activities, these concepts have not been fully embraced by the community. The top five perceived challenges within the lipidomics community were a lack of standardization amongst methods/protocols, lack of lipid standards, software/data handling and quantification, and over-reporting/false positives.
Conclusion
The questionnaire provided an overview of current lipidomics methodologies and further promoted the need for community-accepted guidelines and protocols. The questionnaire also served as a platform to help determine and prioritize metrological issues to be investigated.
Keywords: Lipidomics, Questionnaire, Quantitation, Quality Control, Methodology, Harmonization
1. Introduction
Over the past decade, innovative lipidomic analyses have emerged as an increasingly beneficial strategy for a variety of applications, including those associated with clinical and health biomarker discovery. To date, there are no community-wide established guidelines, protocols, or best practices for lipidomics. A few efforts to harmonize methodologies have been initiated. For example, the 2011 LIPID Metabolites and Pathways Strategy (LIPID MAPS) consortium analysis of Standard Reference Material (SRM) 1950 Metabolites in Frozen Human Plasma (Quehenberger et al. 2010) and the 2017 National Institute of Standards and Technology (NIST) Lipidomics Interlaboratory Comparison Exercise (Bowden et al. 2017a, b) have provided benchmark lipid values to help improve community-wide harmonization efforts. The latter study also helped determine the extent of variability present in lipidomic measurement within the community. Other efforts include proposed lipid nomenclature (Fahy et al. 2005) and shorthand notation of lipids (Liebisch et al. 2013; Koelmel et al. 2017); however, all aspects of the lipidomics workflow warrant consideration for harmonization, including pre-analytical strategies, lipid extraction, analyte separation, mass spectrometric detection, data handling, quality control, quantitation, statistical analysis, biological interpretation, and data dissemination.
We aimed to mirror recent efforts in the metabolomics community (Dunn et al. 2017) by implementing a questionnaire to (1) profile the methodological diversity within a large cohort of the lipidomics community, (2) assess metrological aspects and needs not fully covered in the NIST comparison exercise (e.g., quantitation and quality control), and (3) prioritize future metrological efforts needed in lipidomics.
2. Method information
Between May and August of 2017, NIST conducted a questionnaire within the lipidomics community. Survey invitations were sent to 322 laboratories and advertised at international conferences. In total, 39% of invited laboratories participated (125 in total). The results from the survey are briefly described below, with a more detailed and visual description of all questions and responses provided in the supplemental materials. The data is either presented as a percentage (total number of responses divided by the total number of laboratories responding) or as a total count (number of laboratories that answered with a specific response). Note that for most questions, more than one answer could be selected by each respondent.
The questionnaire information was collected under Office of Management and Budget (OMB) Control #0693–0033—NIST Generic Clearance for Program Evaluation Data Collection and was executed using SurveyMonkey. The survey was comprised of 54 questions, targeting information about the laboratory cohort, lipidomic methodology, quantitation practices, and protocols related to quality control. Most of the questions provided were multiple choice, with the opportunity to select ‘other’ and write-in a different response or explain further.
3. Results and discussion
A major step toward harmonization is the implementation of a workflow that the community can employ to ensure all laboratories achieve the same result (Plebani 2016). Outlined by Tate and Myers (2016), the basic steps toward harmonization generally include (1) instilling community awareness of the need for harmonization, (2) defining the areas of the lipidomics workflow that need harmonization, and (3) engaging the community with activities focused on further defining or ameliorating issues with harmonization. In addition, continuous communication with the community at large during the harmonization process is important to ensure acceptance and implementation of the suggestions. The questionnaire and results described herein attempt to fulfill these steps and address the overall need for community-wide harmonization. More specifically, the questionnaire highlights specific aspects of the lipidomics workflow that need harmonization and reinforces the need to establish community-accepted best practice guidelines.
3.1. Laboratory cohort demographics
The first section of the questionnaire focused on defining the demographics of the lipidomics community (e.g., geographical location, experience, laboratory size, productivity, throughput capacity, and applications). While we aimed to invite the entire lipidomics community to participate, it is important to note that the survey was in English, which may have biased the proportion of participants by country. The response rate for the questionnaire was 39% (125 respondents out of 322 invitations). The 322 invitations were emailed to five continents and 34 countries. The total number of corresponding responses, organized by country, is shown in Supplemental Table S1 and Figure S1. The country with the most responses to the questionnaire was the United States (n = 65; 52%). Of the 125 respondents, 36% were principal investigators. Based on the diverse geographical distribution of participants, it is clear that in addition to lipidomics being utilized in an increasing number of applications and disciplines, lipidomics is also increasingly pervasive worldwide.
A total of 40% of respondents indicated that they are relatively new to the lipidomics community (< 5 years, Supplemental Figure S2). Interestingly, 70% of the laboratories have over five personnel in their laboratory (34% have over 10), which points to the expansion of lipidomic programs within institutions (Supplemental Figure S3). It was also noted that 60% of the respondents have been in the field for over 5 years (n = 75 respondents). Moving forward, it will be critical to engage these experienced lipidomics investigators when the community considers formulating best practice guidelines.
The majority of respondents self-identified as academic entities (74%, Supplemental Figure S4) and pursued applications in the health sciences arena (clinical and medical science, 84%; biomarker discovery, 79%; and drug development/discovery, 24%, Supplemental Figure S5). Other application responses included food science (18%), plant science (14%), natural products (14%), environmental science (10%), toxicology (5%), forensics (1%), and other (19%). Write-in responses for other applications included basic sciences, application development, metabolism and metabolite flux, nutrition, dermatology, and fermentation research.
Beyond experience, we also aimed to define the productivity of current lipidomic laboratories (as defined by number of publications/year and number of samples analyzed/month. A total of 46% of the survey cohort indicated that they publish at least 1–3 manuscripts per year, while 41% reported that they publish more than 3 manuscripts per year (Supplemental Figure S6). Further, 64% indicated that they analyze between 50 and 500 samples per month (Supplemental Figure S7). Of particular note were the laboratories (20%) that indicated that they process over 500 samples per month (68% of these respondents self-identified as academic entities). While harmonized methods and practices are necessary for all lipidomic laboratories, this level of productivity further supports the need to establish guidelines to routinely and transparently assess data quality and comparability.
3.2. Laboratory methodology
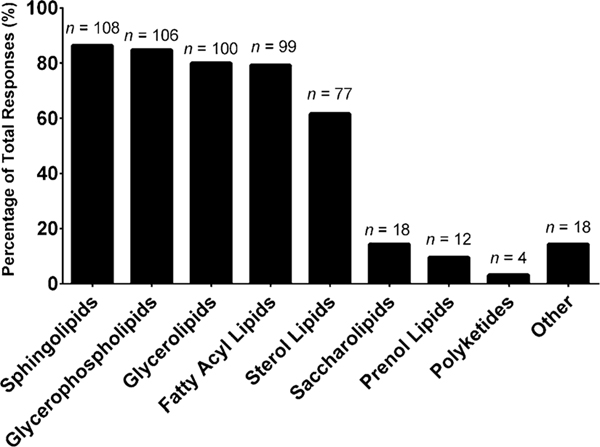
To date, there is no consensus as to the best method to employ for lipidomic experiments. This section of the questionnaire aimed to identify methodological diversity, specifically focusing on the use of internal standards, pre-analytical strategies, sample preparation, analytical platform, lipids analyzed, data processing, lipid annotation, and statistics. Once we establish the diversity of each component of the lipidomics workflow, we can begin to assess the impact of this diversity on lipidomics measurement and subsequently work toward implementing efforts to improve harmonization and standardization within the community. However, before interrogating the diversity in methodology, we aimed to first define what lipids laboratories measured and in what matrices, as both of these aspects can ultimately drive the corresponding methodology employed. The most commonly measured lipid categories, as defined by Fahy et al. (2005), were sphingolipids (86%), glycerophospholipids (85%), glycerolipids (80%), fatty acyl lipids (79%), and sterol lipids (62%), as shown in Fig. 1. The lesser studied lipid categories were saccharolipids (14%), prenol lipids (10%), and polyketides (3%). Interestingly, 14% of the respondents indicated ‘other’ for this question. The write-in responses were largely comprised of lipids representative of the eight major lipid categories (e.g., wax esters, eicosanoids, bile acids), thus suggesting that some laboratories were either unfamiliar with the previously noted lipid category designations or follow a different lipid categorization system.
Fig. 1.
“What lipid categories do you routinely measure in your laboratory (select those that apply)?” The values are shown as a percentage of total responses (total number of laboratories responding, n = 125). The overall total number of responses was 542. The number of responses are indicated in the figure above the solid bars. The x axis labels reflect the survey multiple choice response options. Note that for this question, laboratories had the option to self-identify with more than one option
3.2.1. Sample matrix
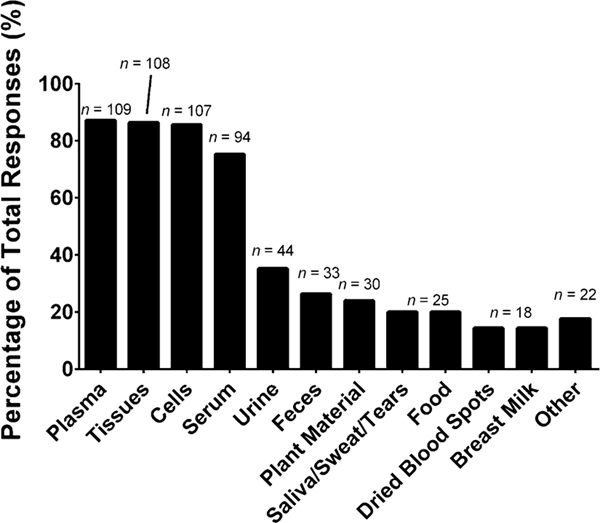
The most commonly employed matrices for lipidomic applications include plasma (87%), tissues (86%), cells (86%), and serum (75%), as shown in Fig. 2. One question we did not ask, but would be worth addressing are the most common types of tissues and cells analyzed as well as the most commonly investigated species. Additional matrices included urine (35%), feces (26%), plant material (24%), saliva/sweat/tears (20%), food (20%), dried blood spots (14%), breast milk (14%), and other (18%). The write-in responses included bronchoalveolar lavage fluid, cerebrospinal fluid (CSF), yeast, microbes, meibum, and model organisms.
Fig. 2.
“What kind of sample matrices does your laboratory analyze for lipidomics (select those that apply)”. The values are shown as a percentage of total responses (total number of laboratories responding, n = 125). The overall total number of responses was 633. The number of responses are indicated in the figure above the solid bars. The x axis labels reflect the survey multiple choice response options. Note that for this question, laboratories had the option to self-identify with more than one option. The write-in responses included bronchoalveolar lavage, cerebrospinal fluid (CSF), yeast, microbes, meibum, and model organisms
3.2.2. Pre-analytical strategies
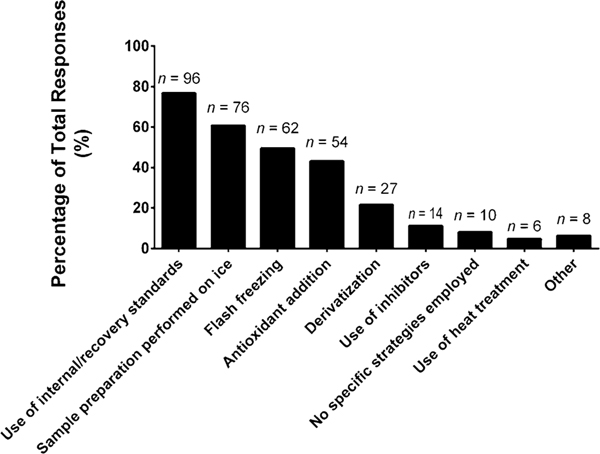
Procedures employed prior to sample extraction, are generally considered the first actionable step in the workflow. We inquired about the types of strategies utilized prior to sample extraction to either enhance or monitor lipid stability (Fig. 3). The most common selection was the use of internal/recovery standards (77%), which is a method aimed at monitoring lipid/sample stability, but has intrinsic limitations. Beyond performing sample preparation procedures on ice (64%), the other common pre-analytical strategies were only employed by half of the respondents (or less) and included flash freezing (50%), antioxidant addition (43%), and use of inhibitors (11%), with the latter two not being applicable to tissue samples. More uncommon, but a potentially promising strategy was heat treatment (5%). It should be noted that methods aimed at enhancing or preserving lipid stability within the lipidomics workflow are not fully vetted.
Fig. 3.
“What strategies (if any) does your laboratory employ for enhancing/monitoring lipid stability (select those that apply)?” The values are shown as a percentage of total responses (total number of laboratories responding, n = 125). The overall total number of responses was 353. The number of responses are indicated in the figure above the solid bars. The x axis labels reflect the survey multiple choice response options. Note that for this question, laboratories had the option to self-identify with more than one option
3.2.3. Sample extraction
The next step in the lipidomics workflow is sample extraction and most labs use one of four extraction procedures, Bligh–Dyer (53%), Folch (43%), methyl tert-butyl ether (MTBE, 38%) and solid phase extraction (30%), as shown in Supplemental Figure S8. MTBE extraction and solid-phase extraction were almost as frequently used as the long-standing traditional chloroform extraction methodologies. Post-extraction, lipid extracts are generally stored by laboratories for over a month (59%, Supplemental Figure S9) and are kept at −80 °C (79%, Supplemental Figure S10).
3.2.4. Analytical instrumentation
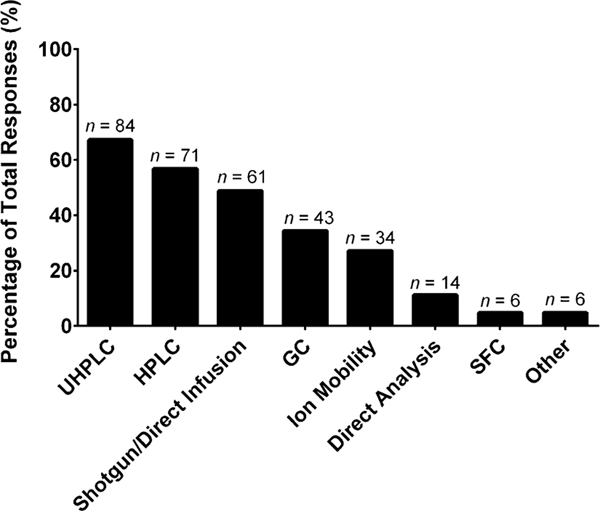
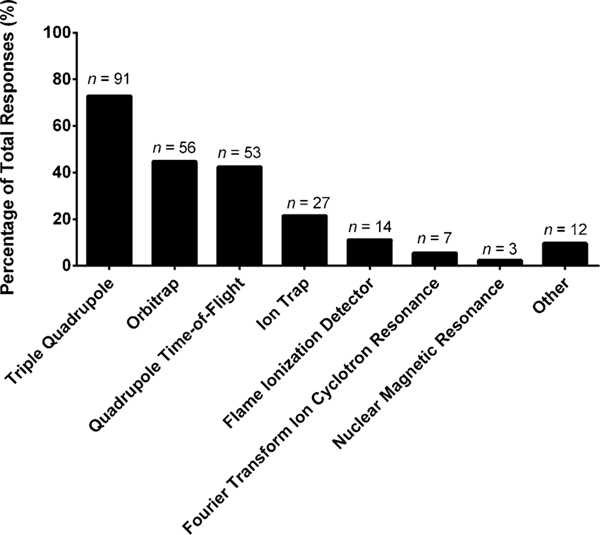
Once a lipid extract is obtained, the extract is introduced (either directly or via chromatography) into an instrument for analysis (e.g., mass spectrometer), various selections are presented in Fig. 4. Most laboratories employ some form of liquid chromatography (ultra-high performance liquid chromatography, UHPLC 67%; high performance liquid chromatography, HPLC 57%) with C-18 columns (58%, optional write-in responses, Supplemental Figure S11). Shotgun lipidomics (49%) and gas chromatography (34%) are still commonly employed. Ion mobility, a newer strategy that is experiencing an uptick in usage for lipidomics, was noted to be employed by 27% of laboratories. The other sample introduction platforms represented were direct analysis (e.g., direct analysis in real time (DART), 11%), supercritical fluid chromatography (5%), and other (5%). Write-in responses included thin-layer chromatography, matrix assisted laser desorption/ionization (MALDI), and imaging. The instrument used to analyze the infused/separated lipid extracts was predominantly mass spectrometry (Fig. 5): triple quadrupole (73%), ion trap (22%), orbitrap (45%), and quadrupole time-of-flight (QTOF, 42%) instruments. Other instruments included the flame ionization detector (11%), Fourier transform ion cyclotron resonance (6%), nuclear magnetic resonance (2%), and other (10%, lesser used platforms noted in Supplemental Material). As an optional follow-up question, if the laboratory incorporated a high-resolution mass spectrometer, we asked what mass resolving power they typically employed to analyze the lipid extracts (Supplemental Figure S12). The most common resolving power intervals were between 26,000 to 50,000 (n = 24) and 51,000 to 100,000 (n = 22). If the respondents are organized by years of experience in the field, in relation to instrument employed, it was found that the newer laboratories (1–5 years) utilized orbitraps 29% and QTOFs 12% more often than the more experienced laboratories (> 5 years), which generally used triple quadrupoles 36%, ion traps 115%, and flame ionization detectors 250% more often than the newer laboratories. This finding may point to a growing transition of the types of workflows employed by laboratories (e.g., targeted to untargeted).
Fig. 4.
“What kind of separation technique does your laboratory use in tandem with mass spectrometry for lipidomics (select those that apply)?” The values are shown as a percentage of total responses (total number of laboratories responding, n = 125). The overall total number of responses was 319. The number of responses are indicated in the figure above the solid bars. The x axis labels reflect the survey multiple choice response options. Note that for this question, laboratories had the option to self-identify with more than one option. Write-in responses included thin-layer chromatography, matrix assisted laser desorption/ionization (MALDI), and imaging
Fig. 5.
“What kind of instruments does your laboratory use for the methods mentioned in Fig. 4 (select those that apply)?” The values are shown as a percentage of total responses (total number of laboratories responding, n = 125). The overall total number of responses was 263. The number of responses are indicated in the figure above the solid bars. The x axis labels reflect the survey multiple choice response options. Note that for this question, laboratories had the option to self-identify with more than one option. Write-in responses included quadrupole and Triple TOF
3.2.5. Data acquisition
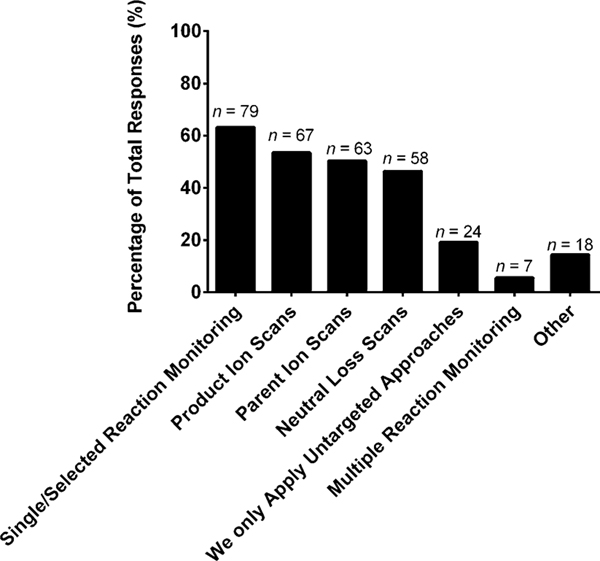
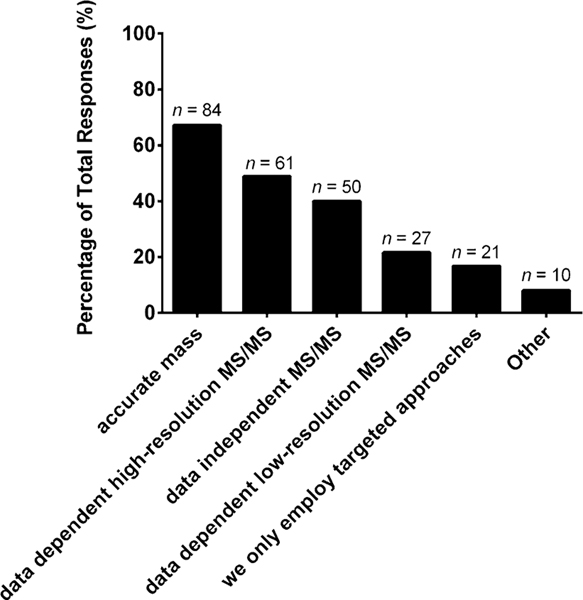
To further investigate the mass spectrometric workflows, we asked what data acquisition methods were incorporated for targeted and untargeted studies. For targeted scanning strategies, single/selected reaction monitoring (63%), product ion scans (54%), parent ion scans (50%), and neutral loss scans (46%) were all used by about half of the participating cohort (Fig. 6). For untargeted workflows, several scanning strategies are commonly employed and included accurate mass (67%), data-dependent high resolution tandem mass spectrometry (49%), and data-independent tandem mass spectrometry (40%), as shown in Fig. 7. The scanning strategies employed for both targeted and untargeted lipidomics workflows were varied, indicating that community-wide protocols need to be established to this level of detail for mass spectrometric operation.
Fig. 6.
“What data acquisition methods does your laboratory incorporate for targeted studies (select those that apply)?” The values are shown as a percentage of total responses (total number of laboratories responding, n = 125). The overall total number of responses was 316. The number of responses are indicated in the figure above the solid bars. The x axis labels reflect the survey multiple choice response options. Note that for this question, laboratories had the option to self-identify with more than one option
Fig. 7.
“What data acquisition methods does your laboratory incorporate for untargeted studies (select those that apply)?” The values are shown as a percentage of total responses (total number of laboratories responding, n = 125). The overall total number of responses was 316. The number of responses are indicated in the figure above the solid bars. The x axis labels reflect the survey multiple choice response options. Note that for this question, laboratories had the option to self-identify with more than one option
3.2.6. Data processing
Perhaps the most methodological diversity was found in data processing, as reported by others (Cajka and Fiehn 2014). Examination of responses, regarding software employed for peak picking/processing (Supplemental Figure S13), indicated that most laboratories (56%) performed this task manually using instrument vendor software (e.g., Xcalibur, MassHunter, MassLynx). It should be noted that over 21 different programs were provided (including the write-in responses). The other software platforms utilized were LipidSearch (21%), XCMS (18%), MZmine (16%), Progenesis QI (14%), Sieve (6%), SimLipid (6%), Compound Discoverer (5%), Lipidyzer (4%), and MS-DIAL (2%). A number of respondents (26%) selected ‘other’ for this question. Upon examination of the write-in responses, the most common answers included MultiQuant (7%) and LipidView (6%), with other lesser responses noted in supplemental materials. Each program has its own algorithms, options, and caveats. Furthermore, the algorithms employed are transparent for some software and proprietary for others. In order to better understand how these programs and options may influence data, the community needs to agree upon the best peak picking/processing workflow(s) and algorithms.
3.2.7. Lipid identification
For lipid identification, the most commonly employed choice was manual examination (59%), as shown in Supplemental Figure S14. It should be noted that over 22 different programs were provided (including write-in responses). The next most commonly employed approach was LipidSearch (30%), followed by LipidBlast (12%). The other programs included Lipidyzer (7%), LipidXplorer (7%), SimLipid (6%), mzCloud (6%), Lipid Data Analyzer (5%), LipidMatch (4%), ALEX (2%), LIMSA (2%), and LipidPioneer (1%). The ‘other’ response (38%) was frequently chosen, and the most common write-in responses were in-house strategies (14%) and LipidView (10%). Other less commonly chosen write-in responses are provided in supplemental materials. New approaches for lipid identification are continuing to be proposed in literature, highlighted by the fact that at least five listed programs were not selected by any respondents. With the plethora of software strategies available, each providing unique features and algorithms, it is important for the community to identify and focus on the most important features and strategies for lipid identification. This is especially important in regards to the large number of researchers who are new to lipidomics, because these researchers may be overwhelmed by the options available and not understand the assumptions and important parameters unique to each software. A well-vetted and community-accepted approach may help lead to a reduction in the over-reporting of lipid annotations, an aspect that was present in our recent interlaboratory study (Bowden et al. 2017a). One aspect that is promising for the harmonization of lipid identification is the fact that most respondents use the LIPID MAPS database (80%), as shown in Supplemental Figure S15. The other databases used for verification included LipidBlast (20%), NIST Mass Spectrometry Database (14%), mzCloud (8%), Japan’s LipidBank (7%), LipidHome (5%), Cyberlipid (5%), SwissLipids (3%), European Lipidomics Initiative (2%), and SphingoMAP (0%, zero respondents selected this option). The other category (26%) included HMBD (6%) and in-house databases (6%).
3.2.8. Lipid annotation
In 2013, Liebisch et al. (2013) suggested a defined set of guidelines for the shorthand annotation of lipids. This paper was critical because it attempted to direct the community toward using an annotation style that was based on the level of structural detail afforded by the methodology and instrumentation employed. The directive was aimed to reduce the over-reporting of structural detail and improve community-wide confidence with lipid identifications. Consequences of not providing lipid annotation to the correct structural detail was recently highlighted by Koelmel et al. (2017), with additional guidelines proposed. Since the paper from Liebisch et al. was proposed almost 5 years ago, we inquired whether the lipidomics community was adopting this proposed style for lipid structures at the fatty acyl level. A total of 69 laboratories responded yes (57%), while 53 laboratories responded no (43%). Based on the software utilized by participants, the percentage who correctly apply this annotation style is most likely significantly less than 57% reported. This highlights that, in addition to individual laboratories, software manufacturers should also accept and implement these guidelines, as many users will assume that annotations obtained are correct. Further, the introduction of new guidelines should also be accompanied with more concrete ways to promote adaptation across the community.
3.2.9. Statistical tools
For statistical analysis, most respondents employed Excel (62%), while all other options were selected less than 35% of the time (Supplemental Figure S15). Additional types of software each laboratory employed for lipid quality control and statistics included R-tools (34%), MetaboAnalyst (22%), GraphPad Prism (18%), MATLAB (18%), SPSS (10%), Origin (6%), PLS_Toolbox (4%), JMP (4%), TraceFinder (4%), Galaxy toolbox (2%), Tableau (2%), Statistica (2%), Minitab (2%), Stata (1%), S-PLUS (1%), Analyze-it (1%), SYSTAT (1%), Orange (0%, zero respondents selected this option), NCSS (0%), and PSPP (0%). The ‘other’ option was selected 37 times (30%). Common write-in responses included Simca (5%) and in-house programs (9%). It should be noted that over 23 different programs were provided (including the write-in responses). There was a great diversity noted in the programs utilized and many of the respondents selected more than one (total selections, n = 281). For improved quantitation and statistics, and subsequent biological interpretation, strategies need to be well-vetted by the community.
3.2.10. Workflow SOPs
Because lipidomic workflows are complex and generally comprise several intricately linked steps, which provide countless opportunities for deviations and missteps, laboratory defined best practices can preserve the quality of the experiment and confident interpretation of obtained data. Standard operating procedures (SOPs), as defined by the International Conference on Harmonization, are “detailed, written instructions to achieve uniformity of the performance of a specific function” (IHT Guideline 1998). Thus, we first asked whether each laboratory employed written SOPs, and if so, what aspects were covered (Supplemental Figure S17). The steps of the lipidomics workflow that received the most responses were sample extraction (78%), instrument calibration/maintenance (66%), and instrument operation (64%). Other components of the lipidomics workflow that had SOPs were sample storage (48%), sample collection (48%), assessment of data quality/quality control (42%), data processing (41%), lipid stability monitoring (20%), and other (8%, e.g., internal standard mix creation). A total of 22 laboratories (18%) indicated that they do not use SOPs in their laboratory. While it may not be critical for certain laboratories to have SOPs, it is clear from these results that implementation of SOPs, across many aspects of lipidomics workflows, needs to be increased at both the individual laboratory level, but also eventually across the entire community. This is especially true for those aspects of the workflow with less than half of the respondents indicating that SOPs exist. For example, the questionnaire showed that strategies aimed to process lipidomic data were extremely diverse. Yet, only 42% of the laboratories have SOPs for this critical process. The implementation and subsequent community-wide dissemination of laboratory SOPs, will lead to the transfer of well-vetted methods to emerging laboratories, promote transparency within the community, and potentially lead to greater comparability among laboratories.
3.3. Lipidomic quantitation
Quantitation is a topic within the lipidomics community that has not been sufficiently harmonized, though there has been some recent literature addressing the topic (Lam et al. 2017; Wang et al. 2016; Yang and Han 2011). A main thrust of the recent interlaboratory comparison exercise for lipidomics was to examine the quantitation of lipids across the community (Bowden et al. 2017a, b). Thus, with this questionnaire, we aimed to investigate methods employed by laboratories for lipid quantitation and address which aspects need community-wide guidelines.
In analytical chemistry, ‘quantitative’ implies that for each compound there is an appropriate internal standard—such that extraction efficiency and matrix effects are fully accounted for when calculating the final quantitative value (Hyötyläinen et al. 2017). This definition is equivalent to absolute quantitation, which is a mode of quantitation largely perceived as a challenge within current lipidomics workflows owing to a lack of commercially available internal standards to cover the lipidome of biological systems. Perhaps noteworthy in this questionnaire is that when asked what type of quantitation was performed in each laboratory (absolute, semi-quant, or relative, as shown in Supplemental Figure S18), 60% of the laboratories noted that they performed some form of absolute quantitation, while only 30% of laboratories reported the use of two or more internal standards on a per lipid class basis (50% of the respondents used only one internal standard). This suggests that many of the laboratories executing an absolute quantitation experiment actually are not. As such, researchers may be under- or overestimating the accuracy of their quantitative measurements. A total of 71 laboratories noted that they perform semi-quantitative lipidomics (57%), while 93 laboratories indicated that they perform relative quantitation (75%). It should also be noted that 92% of the respondents think absolute quantitation is important, and therefore the community is interested in improving quantitation.
The types of internal standards employed were also examined (Supplemental Figure S19). The two most common types of internal standards employed were odd-chain (73%) and deuterated (71%) lipids, largely due to the increased availability of these lipid standards. The lesser employed internal standard types were carbon-13 labeled (29%), low fatty acyl carbon chain (12:0 or less, 24%), isotopic ratio outlier analysis (4%), and other (10%). We investigated whether laboratories made their own internal standard mixtures or purchased them. Results indicated that laboratories typically do both (43%) or exclusively make their own (37%). We also asked the community what lipid classes they thought were the most difficult to quantitate (Supplemental Figure S20). The responses for the lipid classes most difficult to quantitate are shown in decreasing order: phosphatidic acids (32%), eicosanoids (28%), triacylglycerols (19%), phosphatidylinositols (19%), diacylglycerols (15%), free/total fatty acids (14%), phosphatidylserines (15%), cholesterol (13%), lysophosphatidylethanolamines (13%), ceramides (11%), sphingomyelins (11%), lysophosphatidylcholines (10%), cholesteryl esters (10%), phosphatidylethanolamines (9%), bile acids (7%), phosphatidylcholines (7%), phosphatidylglycerols (7%), and ‘other’ (29%). The ‘other’ response had write-ins that included lysophosphatidic acids, vitamins, cardiolipins, steroids, phosphoinositides, glycosphingolipids, plasmalogens, gangliosides, endocannabinoids, monoacylglycerols, cerebrosides, and oxidized lipids. The diverse responses indicate that quantitation is perceived as difficult across the entire lipidome.
We also examined various data processing/handling aspects relevant to lipidomic quantitation, as shown in Supplemental Figure S21. The most commonly employed data handling approach was manual processing (53%). Software programs employed included LipidSearch (21%), Progenesis QI (9%), TraceFinder (6%), Sieve (5%), SimLipid (5%), and Lipidyzer (3%). It should be noted that over 21 different programs were provided (including write-in responses). The most common write-ins were MultiQuant (12%) and MassHunter (3%), with several others briefly mentioned in supplemental material.
One relevant data handling aspect that has not been addressed by the lipidomics community is how to handle multiple adducts (e.g., H+, Na +, NH4+, H−) for an individual lipid species (Supplemental Figure S22). To date, there is no consensus on best practice for this scenario, although based on responses, the most commonly employed approach was to use the most intense adduct for each ionization mode (60%). Other responses that were selected were to report individual adducts (with no further processing, 27%), to sum adducts (20%), or to average adducts (8%). Even individual labs, based on responses, often employ multiple methods, as it was noted that the total answers tallied was 162, indicating that 37 laboratories selected more than one option. A community-wide consensus on the best practice approach for handling multiple adducts (and further multiple LC peaks) per lipid species would be beneficial. The same could be said for using peak height vs peak area (85% of responses indicate the use of the latter).
We also asked each laboratory whether they employed relative response factors (RRFs) for specific lipid categories for quantitation (Supplemental Figure S23). It was found that a majority of responding laboratories do not employ RRFs (64%).
While the question of normalization is clearly application and project dependent, we wanted to examine the diversity associated with this important, yet rarely interrogated aspect of lipid quantitation (Supplemental Figure S24). Clear guidelines are needed for this step, along with a community-wide consensus on the best unit for lipid concentration. The most popular normalization approach was to use total protein (58%). Other popular choices were wet weight (47%), cell count (44%), sum of feature values (which is similar to normalization by TIC, 40%), and dry weight (35%). Some of the lesser used approaches are shown in supplemental information.
3.4. Quality control and reference materials
We aimed to define the prevalence and usage of quality control and reference materials in lipidomic workflows. Quality control samples, an integral component of experimental design and SOPs, are vital tools that provide and/or describe confidence in laboratory procedures and instrument measurement (e.g., technical reproducibility). Based on the survey responses, nearly all laboratories employ quality control measures (Supplemental Figure S25). We asked the respondents what types of QC samples they employed in their laboratory, with the most responses for solvent blanks (71%), pooled samples (matrix-matched, 70%), and extraction blanks (60%). The other QC samples utilized were NIST Standard Reference Materials (SRMs, 21%), pooled samples (not matrix-matched, 20%), and certified reference materials (CRMs, 11%). Six laboratories indicated that they do not use QC samples (5%), while 17 laboratories responded with ‘other’ (14%). The specific QC samples employed by the laboratories (who responded) were mostly in-house materials (n = 58), while commercial lipid mixtures (n = 17), NIST SRMs (n = 15), and other sources were also indicated (n = 8), as shown in Supplemental Figure S26. Most laboratories use QCs, SRMs, or CRMs for technical variance (73%) and method validation (method variance, 69%), as shown in Supplemental Figure S27. Other responses include calibration (52%), trueness of result (35%), metrological traceability (16%), and value assignment of secondary reference materials (6%). Further investigating how laboratories assess the quality of data obtained, we also asked respondents what they used to validate their sample measurements (Supplemental Figure S28). Respondents validate their sample extraction reproducibility with repeated extractions of a sample (74%), instrument reproducibility with repeated instrument analysis of a sample (70%), data quality by reviewing measurements of a previously described quality control sample run in the same batch (51%), and verification of data using either a complementary approach to confirm (17%), a test set (16%), or by sending the analysis to an outside laboratory (3%), and other (3%). Five laboratories (4%) indicated that they do no employ validation activities.
One of NIST’s primary missions is to create/provide useful standards and materials to help improve metrology across disciplines. Thus, we broadly asked what type of reference material would be most desirable to each lipidomic laboratory (Supplemental Figure S29). The responses included a lipid internal standard mixture (70%), lipid standard mixture (54%), spiked standards in a complex biological matrix (47%), and a complex biological matrix (46%). At a more defined level, we asked what types of complex biological reference materials the respondents would most like to see provided (Supplemental Figure S30) and the results were plasma (77%), tissues (66%), serum (54%), cells (50%), urine (30%), plant materials (15%), dried blood spots (15%), saliva/sweat/tears (13%), food (9%), feces (9%), breast milk (6%), and other (7%, e.g., CSF). For those laboratories that do not use commercially available reference materials, we inquired the reason, as shown in Supplemental Figure S31. The most common response was that commercially available reference materials were too expensive (19%), followed by not knowing about them (17%), not having the correct matrix available (16%), and not seeing value in using these materials (9%). Other was selected 26 times (21%) and common write-in responses included difficulties importing these materials into their country, they make their own materials, not large enough quantity per vial, and not suitable for their applications.
3.5. Future of lipidomics
We asked the community what they perceived as the biggest challenge in the lipidomics community, as shown in Supplemental Figure S32. The most common response was a lack of standardization amongst methods/protocols within the community (64%). Other challenges were lack of standards (59%), software/data handling (56%), quantitation (54%), over-reporting/false positives (41%), lipid annotation (37%), and a lack of lipid centric training/workshops (13%). Write-in responses included a lack of established lipid pathways, data sharing, and a lack of reference materials. More specifically for data sharing, 90% of respondents stated that they do not currently store data in a repository (Supplemental Figure S33). Respondents who do store data in repositories listed Metabolomics Workbench, Metabolights, and LIPID MAPS.
We asked if each laboratory has ever participated in an interlaboratory comparison study or ring trial, with 36% of the respondents indicating ‘yes’. This was followed by asking if the laboratories would be interested in participating in a future (NIST) interlaboratory study for lipidomics. The majority responded with ‘yes’ (88%). This implies that the lipidomics community values current harmonization efforts and is engaged. Respondents (73%) felt there were enough opportunities and/or lipidomics conferences per year to present lipidomics studies. Respondents also (97%) would be interested in attending and/or presenting at a proposed Gordon Research Conference focused on the measurement science of lipidomics and metabolomics.
4. Conclusion
4.1. Action items for the lipidomics community
The lipidomics community needs to establish guidelines and best practice protocols to cover all aspects of the lipidomics workflow. This questionnaire serves as a platform to educate the community about current practices, challenges, and opinions regarding lipidomic harmonization. The questionnaire highlights (1) aspects of the lipidomics workflow that need community-wide consensus or further review of best practices, (2) parts of the workflow that need to transition from in-house or manual strategies to more community-accessible methods that the community can assess, analyze, and implement, and (3) areas that need metrological improvement to allow for improved lipid measurement and subsequent community-wide comparability. Below are summarized action items, derived from questionnaire responses, which the community should interrogate further to improve harmonization.
- Develop community best practices/SOPs for both targeted and untargeted lipidomics workflows. Examples include (but are not limited to)**:
- Rigorous studies should investigate lipid stability and pre-analytical strategies. Of special concern is the minimal use of sample stabilization techniques utilized within the community, although lipid degradation is known to occur rapidly.
- Sample extraction, instrument calibration/maintenance, and instrument operation were the most cited lipidomic activities with SOPs. While these aspects of the lipidomics workflow are critical, other important steps should be considered for SOP creation within the community and include sample collection, sample storage, internal standard mix creation, data processing, and assessment of data quality/quality control.
- The lipidomics field should define a set of required metrics and features to validate software programs for data processing, quantitation, statistical analysis, and interpretation of lipidomic data.
Clearly define quantitative approaches (absolute, semi-, and relative) and determine the essential guidelines to perform each approach. Clear guidelines also are needed for the best normalization approach, along with the best quantitation concentration units for each sample type. Commercial outlets, with the guidance of the lipidomics community, need to expand lipid-based standards.
The community needs clear ways to assess the quality of data published. The ability to identify quality lipidomic data sets is a current limitation within the community (Liebisch et al. 2017). One way to achieve this would be to make laboratories report their lipidomic QC data. Subsequently, metrics should be defined to determine the quality of lipidomic data. Laboratories should be also encouraged to share SOPs and methodologies to increase transparency within the community. Another way to better evaluate published lipidomic data, would be to incentivize, or in certain cases mandate laboratories to deposit their lipidomic data into public repositories (only 10% of respondents in this questionnaire currently deposit data into repositories).
Community-wide guidance for incorporating quality control samples into lipidomics workflows would be beneficial. Guidelines pertaining to which QC samples to employ, the number and frequency within a given batch, and performance/acceptability metrics are needed. One example (Bowden et al. 2017a), describes the use of SRM 1950 and community-derived consensus means (and uncertainties) for 339 lipid species, which allow laboratories the ability to extend QC practices beyond their laboratory and compare results with community-wide measurements.
Members of the lipidomics community showed interest in more diverse, low cost, and easily accessible reference materials. Beyond the widely used matrices (e.g., plasma and tissues), an area of recent advancement in the field of disease diagnostics using omics is the application of new, rapidly collected, cost-effective, and minimally intrusive sample matrices. Examples of these emerging sample types include saliva, sebum, and even dried blood spots. The community would benefit from the availability of reference materials for emerging matrices, which would be beneficial for development of new methodologies and quality control for these applications.
Clear and defined guidelines for minimum reporting standards should be implemented by the community, akin to what has been previously proposed by the metabolomics community for chemical and data analysis (Goodacre et al. 2007; Sumner et al. 2007). Furthermore, “White Papers”, similar to the one published by Beger et al. (Beger et al. 2016) should be implemented to provide easily accessible community-wide perspectives regarding relevant issues within lipidomics.
This survey clearly defines a number of challenges in lipidomics. The next step is to establish community-wide best practice guidelines and protocols for performing lipidomic experiments. This could be achieved through future interlaboratory studies, conferences, workshops, training sessions and/or the creation of laboratory networks/focus groups aimed at tackling metrological issues. It should be noted that while we suggest that the laboratories with lipid measurement expertise guide these initiatives, it is imperative that we provide opportunities for all laboratories (regardless of experience) to be engaged and provide feedback.
Supplementary Material
Acknowledgements
This study was partially funded by NIH Grant #U24 DK097209 (R.A.Y.). This research was done in collaboration between Core 1 and Core 3 of the Southeast Center for Metabolomics (SECIM) <http://secim.ufl.edu/>.
Footnotes
Compliance with ethical standards
Conflict of interest All other authors declare no conflicts of interest.
Research involving with human animal participants This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s11306-018-1340-1) contains supplementary material, which is available to authorized users.
Disclaimer Certain commercial equipment or instruments are identified in the paper to specify adequately the experimental procedures. Such identification does not imply recommendations or endorsement by NIST; nor does it imply that the equipment or instruments are the best available for the purpose.
References
- Beger RD, Dunn W, Schmidt MA, Gross SS, Kirwan JA, Cascante M, et al. (2016). Metabolomics enables precision medicine: A white paper, community perspective. Metabolomics, 12, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden JA, Heckert A, Ulmer CZ, Jones CM, Koelmel JP, Abdullah L, et al. (2017a). Harmonizing lipidomics: NIST interlaboratory comparison exercise for lipidomics using standard reference material 1950 metabolites in frozen human plasma. Journal of Lipid Research. 10.1194/jlr.M079012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden JA, Ulmer CZ, Jones CM, & Heckert NA (2017b). Lipid concentrations in standard reference material (SRM) 1950: Results from an interlaboratory comparison exercise for lipidomics. National Institute of Standards and Technology (NIST) IR 8185. http://nvlpubs.nist.gov/nistpubs/ir/2017/NIST.IR.8185.pdf. [Google Scholar]
- Cajka T, & Fiehn O. (2014). Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. Trends in Analytical Chemistry: TRAC, 61, 192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn WB, Broadhurst DI, Edison A, Guillou C, Viant MR, Bearden DW & Beger RD (2017). Quality assurance and quality control processes: Summary of a metabolomics community questionnaire. Metabolomics, 13, 50. [Google Scholar]
- Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Murphy RC, et al. (2005). A comprehensive classification system for lipids. Journal of Lipid Research, 46,, 839–862. [DOI] [PubMed] [Google Scholar]
- Goodacre R, Broadhurst D, Smilde AK, Kristal BS, Baker JD, Beger R. et al. (2007). Proposed minimum reporting standards for data analysis in metabolomics. Metabolomics, 3,, 231–241. [Google Scholar]
- Guideline IHT (1998). ICH Harmonised Tripartite Guideline. International Conference on Hamonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use. Note for Guidance on Statistical Principles for Clinical Trials, ICH Topic E6 (R1), Current step 4 version. [Google Scholar]
- Hyötyläinen T, Ahonen L, Pöhö P, & Orešič M. (2017). Lipidomics in biomedical research-practical considerations. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids, 1862(8), 800–803. [DOI] [PubMed] [Google Scholar]
- Koelmel JP, Ulmer CZ, Jones CM, Yost RA, & Bowden JA (2017). Common cases of improper lipid annotation using high-resolution tandem mass spectrometry data and corresponding limitations in biological interpretation. Biochimica et Biophysica Acta (BBA): Molecular and Cell Biology of Lipids, 1862(8), 766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SM, Tian H, & Shui G. (2017). Lipidomics, en route to accurate quantitation. Biochimica et Biophysica Acta (BBA): Molecular and Cell Biology of Lipids, 1862(8), 752–761. [DOI] [PubMed] [Google Scholar]
- Liebisch G, Ekroos K, Hermansson M, & Ejsing CS (2017). Reporting of lipidomics data should be standardized. Biochimica et Biophysica Acta (BBA): Molecular and Cell Biology of Lipids, 1862, 747–751. [DOI] [PubMed] [Google Scholar]
- Liebisch G, Vizcaíno JA, Köfeler H, Trötzmüller M, Griffiths WJ, Schmitz G, et al. (2013). Shorthand notation for lipid structures derived from mass spectrometry. Journal of Lipid Research, 54, 1523–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plebani M. (2016). Harmonization of clinical laboratory information: Current and future strategies. EJIFCC, 27, 15–22. [PMC free article] [PubMed] [Google Scholar]
- Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, et al. (2010). Lipidomics reveals a remarkable diversity of lipids in human plasma. Journal of Lipid Research, 51, 3299–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, et al. (2007). Proposed minimum reporting standards for chemical analysis. Metabolomics, 3, 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JR, & Myers GL (2016). Harmonization of clinical laboratory test results. EJIFCC, 27, 5–14. [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wang C, & Han X. (2016). Selection of internal standards for accurate quantification of complex lipid species in biological extracts by electrospray ionization mass spectrometry: What, how and why? Mass Spectrometry Reviews, 36(6), 693–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, & Han X. (2011). Accurate quantification of lipid species by electrospray ionization mass spectrometry—meets a key challenge in lipidomics. Metabolites, 1, 21–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.