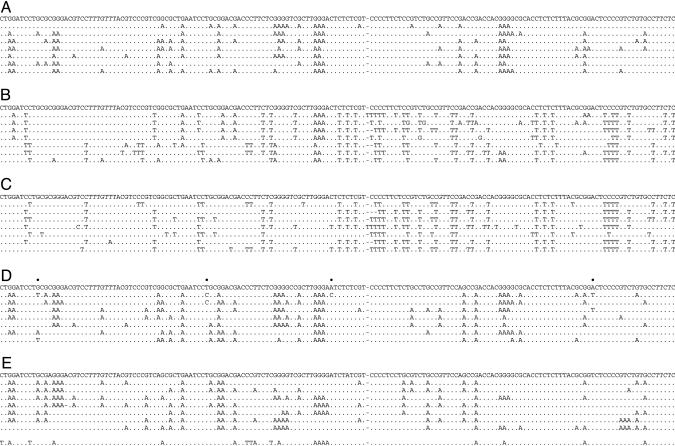
Fig. 2.
Massive deamination of HBV DNA by APOBEC3 cytidine deaminases. A selection of hypermutated HBV sequences mapping just 5′ to ORF X are shown compared with their reference. The sequence is given with respect to the viral plus strand. Only differences are shown. Hyphens denote gaps introduced to maximize sequence identity. All sequences were unique, indicating that they corresponded to distinct molecular events. (A) Selection of G → A hypermutations resulting from APOBEC3G deamination of minus-strand DNA. (B) A selection of mixed G → A and C → T hypermutants from APOBEC3B, -3F, and -3G cotransfections. (C) A selection of uniquely C → T hypermutants from APOBEC3B, -3F, and -3G cotransfections. (D and E) Hypermutated sequences derived from virions in the sera of patients 130.71 and 12763, respectively. The reference sequence given was the major sequence among 10 derived from PCR products amplified at 95°C. Dots above the reference sequences indicate polymorphic sites among the 10 nonhypermutated sequences. For patient 12763 (Fig. 3E), one of the sequences was a mixed G → A + C → T hypermutant.