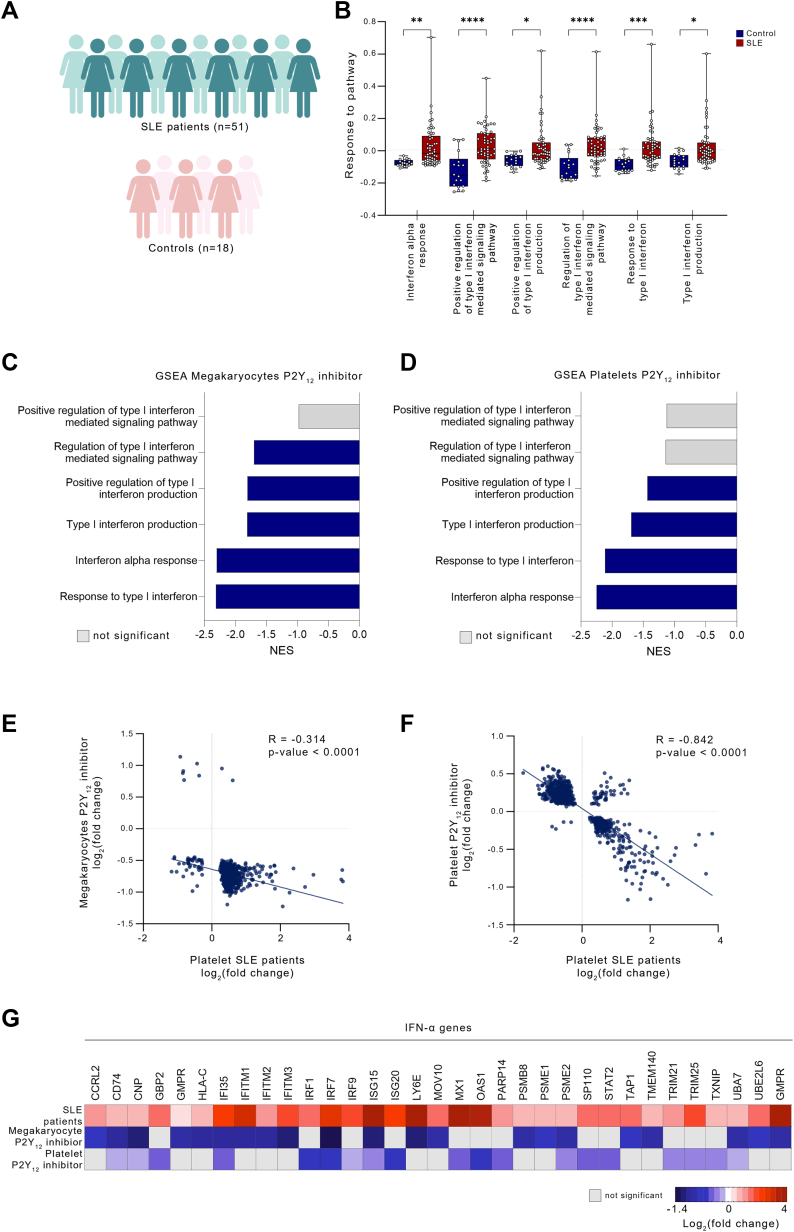
Figure 3.
P2Y12 Inhibition Attenuates IFN Genes Up-Regulated in Patients With SLE
(A) Experimental workflow: Platelets were collected from patients with systemic lupus erythematosus (SLE) and healthy control subjects, RNA was isolated, and RNA-sequencing was performed. (B) Hallmark and Gene Ontology pathway analysis, focused on IFNα and type I IFN in platelets from patients with SLE (P < 0.05). Data presented as mean ± SD; P values were determined from the gene set enrichment analysis (GSEA) output. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. (C, D) Overlapping pathways in P2Y12-inhibited MKs and platelets (blue = P < 0.05; gray = P = NS). (E, F). Scatterplots of fold change differences comparing platelets of patients with SLE vs control subjects and P2Y12-inhibited MKs (E) and platelets (F); P < 0.05. (G) Heatmap of differentially expressed genes in platelets from patients with SLE and P2Y12-inhibited MKs and platelets. The genes represent IFNα-related genes, up-regulated in patients with SLE, and down-regulated on P2Y12 inhibition in MK and platelet groups. Abbreviations as in Figure 1.