Abstract
A comprehensive three-dimensional digital brain atlas of cortical and subcortical regions based on MRI and histology has a broad array of applications in anatomical, functional, and clinical studies. We first generated a Subcortical Atlas of the Marmoset, called the “SAM,” from 251 delineated subcortical regions (e.g. thalamic subregions, etc.) derived from high-resolution Mean Apparent Propagator-MRI, T2W, and magnetization transfer ratio images ex vivo. We then confirmed the location and borders of these segmented regions in the MRI data using matched histological sections with multiple stains obtained from the same specimen. Finally, we estimated and confirmed the atlas-based areal boundaries of subcortical regions by registering this ex vivo atlas template to in vivo T1- or T2W MRI datasets of different age groups (single vs. multisubject population-based marmoset control adults) using a novel pipeline developed within Analysis of Functional NeuroImages software. Tracing and validating these important deep brain structures in 3D will improve neurosurgical planning, anatomical tract tracer injections, navigation of deep brain stimulation probes, functional MRI and brain connectivity studies, and our understanding of brain structure–function relationships. This new ex vivo template and atlas are available as volumes in standard NIFTI and GIFTI file formats and are intended for use as a reference standard for marmoset brain research.
Keywords: subcortical atlas, MAP-MRI, histology, SAM, marmoset monkey
Introduction
The basal ganglia, thalamus, hypothalamus, brainstem nuclei, and amygdala are subcortical regions that regulate sensorimotor, cognitive, limbic, and autonomic (sympathetic and parasympathetic) functions (Lanciego et al. 2012; Ferrazzoli et al. 2018; Sklerov et al. 2019; Aggleton and O'Mara 2022). A comprehensive 3D digital template atlas of these subcortical regions in non-human primates (NHPs) based on MRI and histology is of great value in anatomical, functional, and clinical studies. In particular, a 3D atlas can be registered to an NHP brain MRI volume to best determine the regions of interest (ROIs) for anatomical tracer injections for connectome studies, the areal location of observed functional MRI (fMRI) responses, and the potential targets for deep brain stimulation (DBS) in NHP models of neurological disorders. In this study, we developed a Subcortical Atlas of the Marmoset (SAM) monkey using ultra-high-resolution Mean Apparent Propagator (MAP)-MRI, T2W, and magnetization transfer ratio (MTR) images combined with a corresponding set of histological images from the same marmoset brain. The common marmoset (Callithrix jacchus) is a useful NHP model for aging studies (Tardif et al. 2011; Okano et al. 2016) and has cytoarchitectonic brain areas similar to those of macaques (Burman and Rosa 2009; Fukushima et al. 2019). The early stages of sensory and motor processing in the marmoset are similar to other primates (Kaas 2020). Thus, the results from this NHP species are relevant for making inferences about how sensorimotor and other systems are organized and function in humans.
MAP-MRI (Ozarslan et al. 2013) provides a comprehensive and clinically feasible (Avram et al. 2016) assessment of water diffusion in tissues. In each voxel, MAP-MRI explicitly measures the probability density function of the net 3D microscopic displacements of diffusing water molecules, also known as diffusion propagators. MAP-MRI subsumes and generalizes other diffusion MRI (dMRI) signal representations (Avram et al. 2017) and quantifies average diffusion properties in isotropic and anisotropic tissues with arbitrary microstructure and architecture thoroughly and accurately with multiple microstructural markers. Besides the diffusion tensor imaging (DTI)-derived fractional anisotropy (FA), axial, radial, and mean diffusivities (AD, RD, and MD, respectively), and direction-encoded color (DEC) maps (Pajevic and Pierpaoli 1999), MAP-MRI yields a family of new microstructural parameters that capture more subtle features of diffusion propagators, such as zero-displacement probabilities, non-gaussianity index (NG), and propagator anisotropy (PA) (Ozarslan et al. 2013; Avram et al. 2014a, 2017, 2018a), as well as MAP-derived orientation distribution functions. Taken together, the DTI/MAP parameters provide a more sensitive and specific microstructural assessment compared with conventional dMRI (Hutchinson et al. 2018) and structural (e.g. T1W and T2W) scans. They have proven remarkably effective for detailed anatomical segmentation of the cortical (Avram et al. 2022b) and subcortical structures (Saleem et al. 2021, 2023).
Many studies have segmented subcortical structures and provided 3D atlases in humans using high-resolution in vivo MRI with multiple image contrasts or ex vivo spin echo (SE) T2W MRI with histological stains (Rijkers et al. 2007; Abosch et al. 2010; Lenglet et al. 2012; Deistung et al. 2013a, 2013b; Keuken et al. 2014; Ewert et al. 2018; Pauli et al. 2018; Hoch et al. 2019a, 2019b). In contrast, only a limited number of studies have provided detailed maps of subcortical regions or created a 3D digital subcortical atlas template using combined MRI and histology in the marmoset. (i) Newman and colleagues described a subcortical atlas of a marmoset monkey (Newman et al. 2009). However, it is a non-digital version based on a series of individual photographs of Nissl sections with closely matched MR images obtained from a different marmoset brain. A similar type of histology-based atlases in book form has been published with or without MRI (Stephan et al. 1980; Palazzi and Bordier 2008; Yuasa et al. 2010; Hardman and Ashwell 2012; Paxinos et al. 2012; Iriki et al. 2018). (ii) The Brain/Minds digital-marmoset brain atlas has provided 3D segmentation of major cortical and subcortical regions using a standard structural T1/T2W MRI, coregistered with only Nissl-stained histology data to identify the ROI (Hashikawa et al. 2015; Woodward et al. 2018). (iii) Liu and colleagues (Liu et al. 2018) constructed a 3D digital marmoset brain atlas on ex vivo MTR, T2W, and dMRI. This study provided the segmentation of 54 cortical, but few subcortical (n = 16) regions in their atlas, and the area labels were derived from a different subject (Paxinos et al. 2012). (iv) A recent study generated a histology-based probabilistic atlas of the marmoset brain called the Nencki–Monash template, but it focused only on the cytoarchitectonic parcellation of cortical areas (Majka et al. 2020, 2021). Similar to the marmoset, only a few studies have produced detailed mapping of subcortical regions in the macaque monkey using combined MRI and histology (for more details, see the introduction section in Saleem et al. 2021).
Many of the subcortical nuclei and their subregions are challenging to identify and delineate in conventional MRI because of their small size, buried location, and often subtle contrast compared with neighboring regions. As shown in our previous study in the macaque (Saleem et al. 2021) and marmoset (Saleem et al. 2023) monkeys, combining volumes of different MRI markers acquired with high-spatial resolution (100–200 μm), aided by whole-brain histological information derived from the same brain specimen, is key to delineating nuclei and fiber tracts in deep brain structures, including substructures and laminae, e.g. in the thalamus and basal ganglia. Thus, segmentation and validation of brain areas based on MRI-histology correlations are crucial for constructing accurate 3D digital template atlases in NHPs.
In this study, we combined MAP-MRI, T2W, and MTR data with matched high-resolution images of histological sections with multiple stains derived from the same ex vivo marmoset brain specimen to segment 251 subcortical regions and associated white matter pathways. This integrated multimodal approach yields a more objective and reproducible delineation of gray matter nuclei and their boundaries in deep brain structures, including the basal ganglia, thalamus, hypothalamus, limbic region (amygdala), basal forebrain, and rostrocaudal extent of the brainstem (midbrain, pons, and medulla). We derived the 3D SAM brain from these segmented deep brain regions and registered this volume to a range of in vivo T1- and T2W standard MRI volumes from control subjects in different age groups to illustrate the application of the atlas to in vivo studies. This newly derived ex vivo 3D digital atlas is intended to provide a practical standard template for neuroanatomical, functional (fMRI), clinical, and connectional imaging studies involving subcortical targets in marmoset monkeys.
Materials and methods
Perfusion fixation
One adult male marmoset monkey (C. jacchus), weighing 340 g, was used in this study. This animal was previously involved in transgenic studies at the NIMH/NIH transgenic core, and we received a perfusion-fixed brain specimen of this animal from this core for our ex vivo MRI and histological studies. All procedures adhered to the Guide for the Care and Use of Laboratory Animals (National Research Council) and were performed under a protocol approved by the Institutional Animal Care and Use Committee of the National Institute of Mental Health (NIMH) and the National Institutes of Health (NIH). The animal was deeply anesthetized with sodium pentobarbital and perfused transcardially with 0.5 l of heparinized saline, followed by 2 l of 4% paraformaldehyde, both in 0.1 M phosphate buffer (pH 7.4). After perfusion, the brain was removed from the cranium, photographed, and post-fixed for 24 h in the same buffered paraformaldehyde solution and then transferred into 0.1 M phosphate-buffered saline with sodium azide before MRI scanning.
Ex vivo MRI
Data acquisition
The MRI data acquisition and processing were described in detail in our previous study (Saleem et al. 2023). In brief, we positioned the fixed marmoset brain specimen in a 3D printed brain mold and then inside a custom 30 mm diameter cylindrical container. We then filled the 3D mold and container with Fomblin and prepared the sample for MR imaging using a Bruker 7 T/300 mm horizontal MRI scanner with a 30 mm inner diameter quadrature millipede coil (ExtendMR; http://www.extendmr.com/).
We acquired MAP-MRI data using a diffusion spin-echo (SE) 3D echo-planar imaging (EPI) sequence using the following imaging parameters: 150 μm isotropic resolution, 4.32 × 2.76 × 2.76 cm3 field-of-view (FOV), 288 × 184 × 184 imaging matrix, 10 segments per kz-plane, and 1.33 partial Fourier acceleration. We acquired 256 diffusion-weighted images (DWIs) with diffusion gradient pulse durations and separations of δ = 6 ms and Δ = 28 ms, respectively, a 48 ms echo time (TE) and a 650 ms repetition time (TR). MAP-DWIs were acquired using multiple b-value shells: 100, 500, 1000, 1500, 2500, 3500, 4500, 5250, 7000, 8500, 10,000 s/mm2 with uniformly sampled gradient directions (Koay et al. 2012; Avram et al. 2018b) on each shell (7, 10, 12, 15, 21, 22, 27, 31, 35, 36, and 40, respectively). We also obtained a magnetization transfer (MT) prepared scan using a 3D gradient echo sequence with 150 μm isotropic resolution (288 × 184 × 184 imaging matrix, 4.32 × 2.76 × 2.76 cm FOV with a 15° excitation flip angle, TE/TR = 3.7/37 ms, 2 averages, MT saturation pulse with 2 kHz offset, 12.5 ms Gaussian pulse with 6.74 μT peak amplitude, and 540° flip angle). The total duration of the MAP-MRI scan was 85 h and 20 min, and the MT scan was 11 h and 8 min.
Data processing
From the scans acquired with and without MT, we computed the MT ratio (MTR) volume with good GM/WM tissue contrast to serve as a structural template for subsequent DWI distortion correction and co-registration. We processed the MAP-MRI dataset with the comprehensive Tortoise pipeline (Pierpaoli et al. 2010) that performs denoising, corrects for Gibbs ringing, gradient eddy currents, intra-scan drift, and EPI distortions, and registers all volumes to the MTR scan. We estimated diffusion propagators by fitting the data in each voxel with a MAP series truncated at order 4 and computed DTI (MD, AD, RD); FA (fractional anisotropy); CL, CP, CS-linear, planar, and spherical anisotropy coefficients, respectively (Westin et al. 2002) and MAP-MRI tissue parameters (RTOP-return-to-origin probability, RTAP-return-to-axis probability, RTPP-return-to-plane probability, PA-propagator anisotropy, and NG), along with the corresponding baseline (T2W) volume. Furthermore, we estimated and visualized the fiber orientation distribution functions (FODs) using MRtrix 3.0.1 (Tournier et al. 2012). For more details, see our previous study (Saleem et al. 2023).
Histological processing and data analysis
All histological procedures (section cutting and staining) are described in detail in Saleem et al. (2023). Following MRI acquisition, we prepared the whole-brain specimen as one tissue block for histological processing using 5 different stains. Serial frozen coronal sections (50 μm thick) were cut on a sliding microtome through the entire brain, including the cerebrum, brainstem, and cerebellum. We collected 670 brain sections, sorted them into 5 parallel series (134 sections per set with 250 μm spacing between adjacent sections), and sequentially stained them for parvalbumin (PV), neurofilament (SMI 32), acetylcholinesterase (AchE), Cresyl violet (CV), and choline acetyltransferase (ChAT) (Fig. 1). We stained all the sections in this animal and collected block-face images of a frozen tissue block at every 250 μm interval. The commercially available antibodies used for SMI-32, PV, and ChAT staining are indicated in the “Key Resources Table (Table 2).”
Fig. 1.
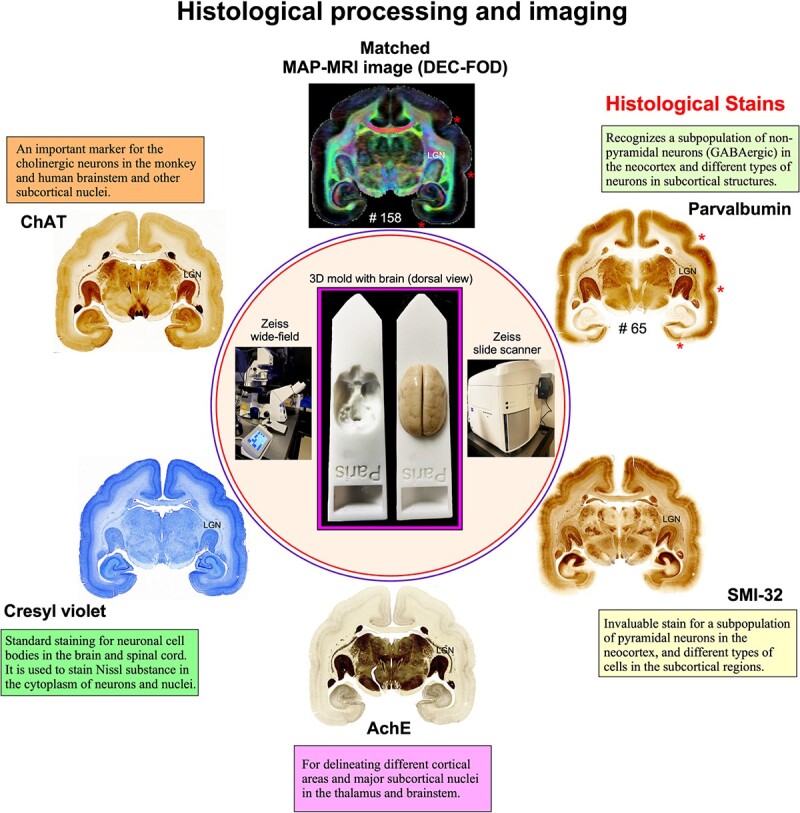
Histological staining and high-resolution imaging. Frozen sections were cut coronally from the frontal cortex to the occipital cortex at 50 μm thickness on a sliding microtome. In total, 670 sections were collected, and all sections were processed with different cell bodies and fiber stains. This example shows an adjacent series of 5 sections at the level of the anterior temporal cortex stained with PV, SMI-32, AchE, CV, and ChAT. The unique characteristics of each stain are described in the “Histological markers” in Results section. We obtained 134 stained sections in each series, and the interval between 2 adjacent sections in each series is 250 μm. High-resolution images of stained sections were captured using a Zeiss wide-field microscope and a Zeiss Axioscan Z1 high-resolution slide scanner (inset). These histological images were then aligned manually with the corresponding MAP-MRI (top) and other MRI parameters of the same specimen to allow visualization and delineation of subcortical structures in a specific ROI. In both MRI and histology sections, note the correspondence of sulci (“stars” on the top right), gyri, and deep brain structures (e.g. LGN). #65 refers to the section number in each set/series of stained sections, and #158 indicates the matched MRI slice number in 3D volume. The inset shows the 3D brain mold with and without the brain specimen for MRI.
Table 2.
Key resources (atlas data, antibodies for histological staining, and software).
| Online resource (MRI and Histology) | References | Identifier (Online links) |
|---|---|---|
| MRI and Atlas data | This paper | |
| MRI templates and SAM Atlas with area labels | https://afni.nimh.nih.gov/pub/dist/atlases/marmoset/SAM_Marmoset/SAM_marmoset_subcortical_dist.tgz | |
| Table 1 with delineated subcortical areas | ||
| Histology-Immunohistochemistry (antibodies) | Saleem et al. (2023) | |
| Anti-nonphosphorylated neurofilament H | https://www.biolegend.com/ | |
| (clone SMI-32, Cat # 801701 | ||
| Anti-Parvalbumin antibody (Cat. # P3088) | https://www.sigmaaldrich.com/ | |
| Anti-choline acetyltransferase antibody (Cat. # AB144P) | https://www.sigmaaldrich.com/ | |
| Software | ||
| ITK-SNAP version 4.0 | Yushkevich et al. (2006) | http://www.itksnap.org/pmwiki/pmwiki.php |
| Mrtrix 3.0.1 | Tournier et al. (2012) | https://www.mrtrix.org/ |
| Canvas X Draw 7.0.3 | Saleem et al. (2021, 2023) | https://www.canvasgfx.com/products/canvas-x-draw |
| Adobe Photoshop version 24.2 | Saleem et al. (2021, 2023) | https://www.adobe.com/ |
| AFNI Version 22.1.10 | Cox (1996) | https://afni.nimh.nih.gov/ |
Histological data analysis
High-resolution images of all stained sections were captured using a Zeiss wide-field microscope and a Zeiss high-resolution Axioscan-Z1 slide scanner at 5× objective, and these digital images were adjusted for brightness and contrast using Adobe Photoshop (v24.2). These images were then aligned manually with the corresponding images of DTI/MAP parameters along with the estimated T2W (i.e. non-diffusion weighted) and MTR images to allow visualization and delineation of subcortical structures in specific ROIs (e.g. Figs 3 and 4). Some structures, such as the striatum and pallidum, were demarcated by juxtaposing matched MRI and histology sections, but for others (e.g. amygdala), we used a different approach to outline their subregions, as described in our previous study (Saleem et al. 2021). In brief, we first superimposed a histological section onto the matched MRI slice and then manually rotated and proportionally scaled it to match the boundaries of the different nuclei and their subregions on the MRI using the transparency function in Canvas X Draw software 7.0.3. Finally, the borders of the subregions were manually traced on the histology sections and translated onto the superimposed underlying MR images using the polygon-drawing tools and smooth and grouping functions in this software (Saleem et al. 2021, 2023).
Fig. 3.
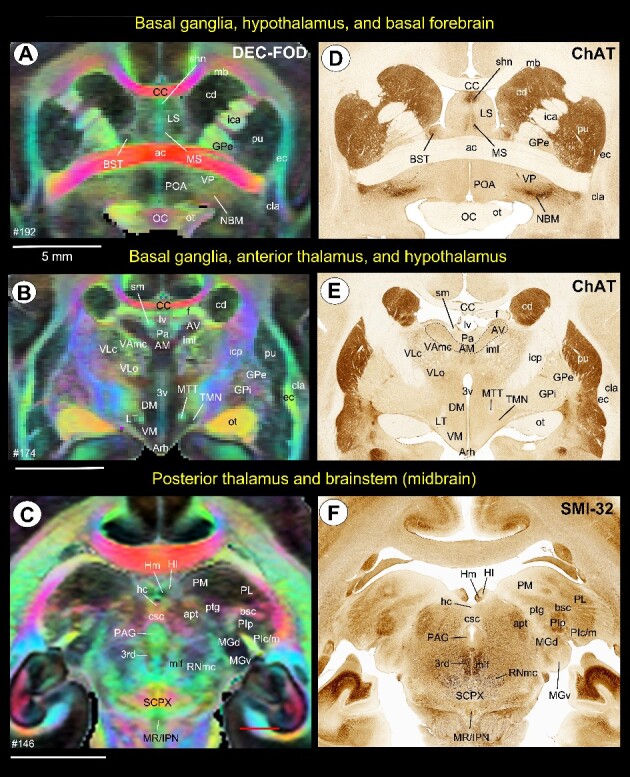
Subcortical areas for the 3D atlas (SAM). (A–F) Examples showing the basal ganglia, thalamus, hypothalamus, basal forebrain, and brainstem that are identified and segmented on the MAP-MRI (DEC-FOD) with reference to matched histological sections stained with ChAT and SMI-32, and other stained sections (not shown here). Abbreviations: 3rd, third cranial (oculomotor) nuclei; 3v, 3rd ventricle; ac, anterior commissure; AM, anterior medial nucleus; apt, anterior pretectal nucleus; Arh, arcuate hypothalamic nucleus; AV, anterior ventral nucleus; bsc, brachium of superior colliculus; BST, bed nucleus of stria terminals; CC, corpus callosum; cd, caudate nucleus; cla, claustrum; csc, commissure of superior colliculus; DM, dorsomedial hypothalamic area; ec, external capsule; f, fornix; GPe, globus pallidus, external segment; GPi, globus pallidus, internal segment; hc, habenular commissure; Hl, lateral habenular nucleus; Hm, medial habenular nucleus; ica, internal capsule, anterior limb; icp, internal capsule, posterior limb; iml, internal medullary lamina; IPN, interpeduncular nucleus; LS, lateral septum; LT, lateral hypothalamic area; lv, lateral ventricle; mb, Muratoff bundle; MGd, medial geniculate nucleus, dorsal division; MGv, medial geniculate nucleus, ventral division; mlf, medial longitudinal fasciculus; MR, median raphe; MS, medial septum; MTT, mammillothalamic tract; NBM, nucleus basalis of Myenert; oc, optic chiasm; ot, optic tract; Pa, paraventricular nucleus; PAG, periaqueductal gray; PIc, inferior pulvinar, caudal division; PIm, inferior pulvinar, medial division; PIp, inferior pulvinar, posterior division; PL, lateral pulvinar; PM, medial pulvinar; POA, preoptic area; ptg, posterior thalamic group; pu, putamen; RNmc, red nucleus, magnocellular division; SCPX, superior cerebellar peduncle decussation; shn, septo-hippocampal nucleus; Sm, stria medullaris; TMN, tuberomammillary nucleus; VAmc, ventral anterior nucleus, magnocellular division; VLc, ventral lateral caudal nucleus; VLo, ventral lateral oral nucleus; VM, ventromedial hypothalamic area; VP, ventral pallidum. Scale bars: 5 mm applies to A–F.
Fig. 4.
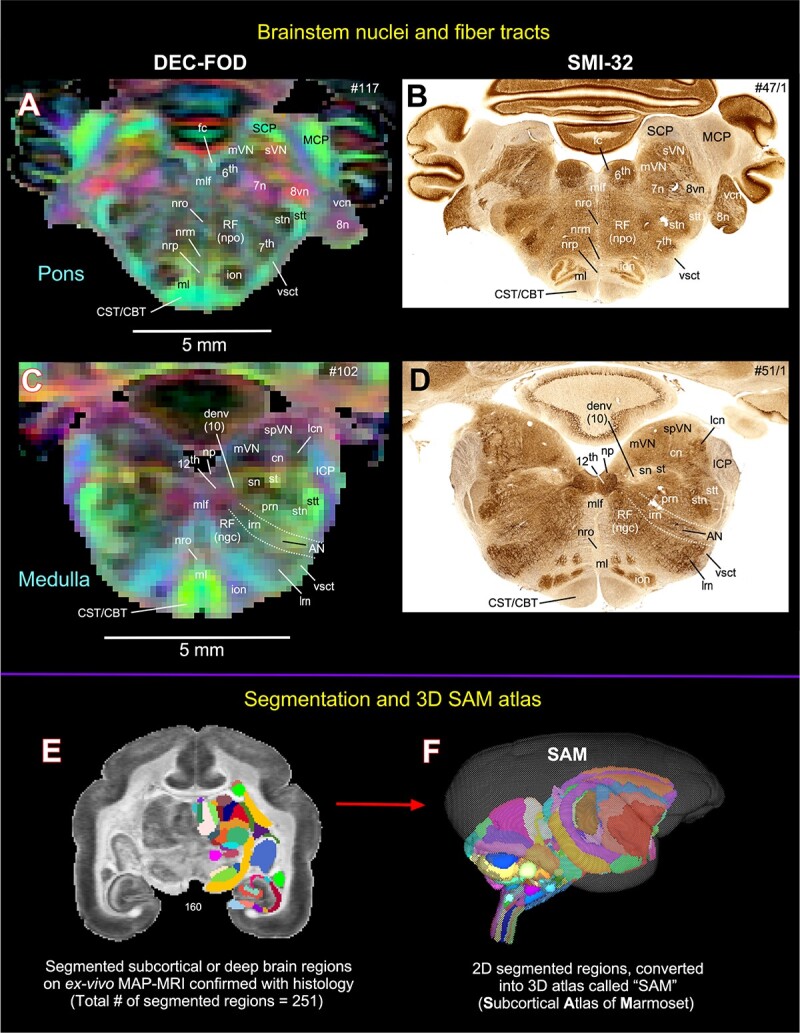
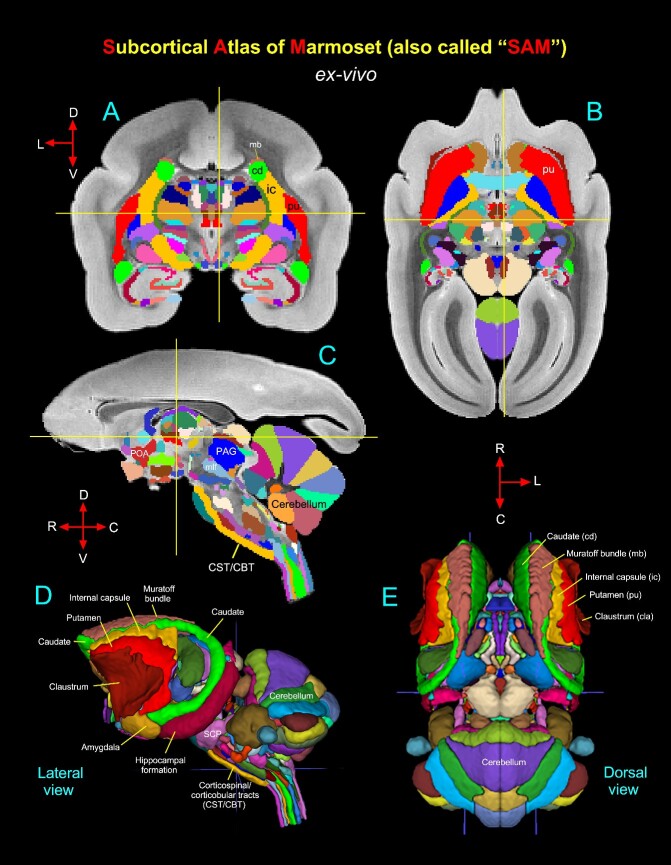
Subcortical areas for the 3D atlas (SAM). (A–D) More examples show the subcortical areas at the brainstem level (pons and medulla) that are identified and segmented on MAP-MRI (DEC-FOD) with reference to matched histological sections stained with SMI-32, and other stained sections (not shown here). Abbreviations: 6th, abducent nuclei; 7th, facial nuclei; 7n, facial nerve; 8n, vestibulocochlear nerve; 8vn, vestibular nerve; 12th, hypoglossal nucleus; AN, ambiguous nucleus; CBT, corticobulbar tract; cn, cuneate nucleus; CST, corticospinal tract; denv (10), dorsal motor nucleus of vagus; fc, facial colliculus; ICP, inferior cerebellar peduncle; ion, inferior olivary nucleus; irn, intermediate reticular nucleus; lcn, lateral cuneate nucleus; lrn, lateral reticular nucleus; MCP, middle cerebellar peduncle; ml, medial lemniscus; mlf, medial longitudinal fasciculus; mVN, medial vestibular nucleus; np, nucleus prepositus; nrm, nucleus raphe magnus; nro, nucleus raphe obscurus; nrp, nucleus raphe pallidus; prn, parvicelluar reticular nucleus; RF (ngc), reticular formation, nucleus gigantocellularis; RF (npo), reticular formation, nucleus pontis centralis oralis; SCP, superior cerebellar peduncle; sn, solitary nucleus; spVN, spinal vestibular nucleus; st, solitary tract; stn, spinal trigeminal nucleus; stt, spinal trigeminal tract; sVN, superior vestibular nucleus; vcn, ventral cochlear nucleus; vsct, ventral spinocerebellar tract. Subcortical segmentation and 3D ex vivo digital template atlas. (E) Two hundred and fifty-one deep brain regions, including the HF and cerebellum, were manually segmented through a series of 150 μm thick MAP-MRI sections using ITK-SNAP. (F) A 3D isosurface rendering of the individual regions within a volume rendering of the T2W dataset. This new MRI-histology-based segmented volume (called ex vivo “SAM”) is registered to an in vivo multi-subject population-based T1W MRI volume oriented to the EBZ stereotaxic coordinate system (Liu et al. 2021) or a range of in vivo T2W MRI volumes of marmoset monkeys with different age groups and genders. For more details, see Figs 6 and 7.
The histological sections matched well with the MRIs in this study. It was not required to resample the MRI volume to achieve good alignment with the histological images in the current study. Alignment of these images was only possible by carefully orienting the entire brain specimen with reference to sagittal MR images from this specimen. These steps were performed on the microtome stage before attaching and freezing the brain specimen with dry ice. These steps enabled us to match the sulci, gyri, and ROI in deep brain structures in both MRI and histological sections, as shown in Fig. 1 and the Results section below (Figs. 3 and 4).
Delineation of subcortical regions and generation of standard “SAM” (ex-vivo) atlas
Subregions (ROIs) of the basal ganglia, thalamus, hypothalamus, amygdala, brainstem, and other deep brain areas and selected fiber bundles were manually segmented through a series of 146, 150 μm thick coronal sections in PA/DEC, T2W, or other MRI parameters using ITK-SNAP (v4.0) (Yushkevich et al. 2006). The spatial extent and borders of each segmented region in MRI were confirmed using matched high-resolution histology images obtained from multiple stained sections (Figs. 1 and 2). The regions were drawn manually only on the left hemisphere. To define a good axis of symmetry, the MRI dataset was rotated about the anterior–posterior axis by a small angle of 0.75°in Analysis of Functional NeuroImages (AFNI)’s Nudge plugin. The dataset coordinates were aligned to an Anterior Commissure—Posterior Commissure (AC-PC) axis with the center of the AC at the (0,0,0) XYZ coordinate.
Fig. 2.
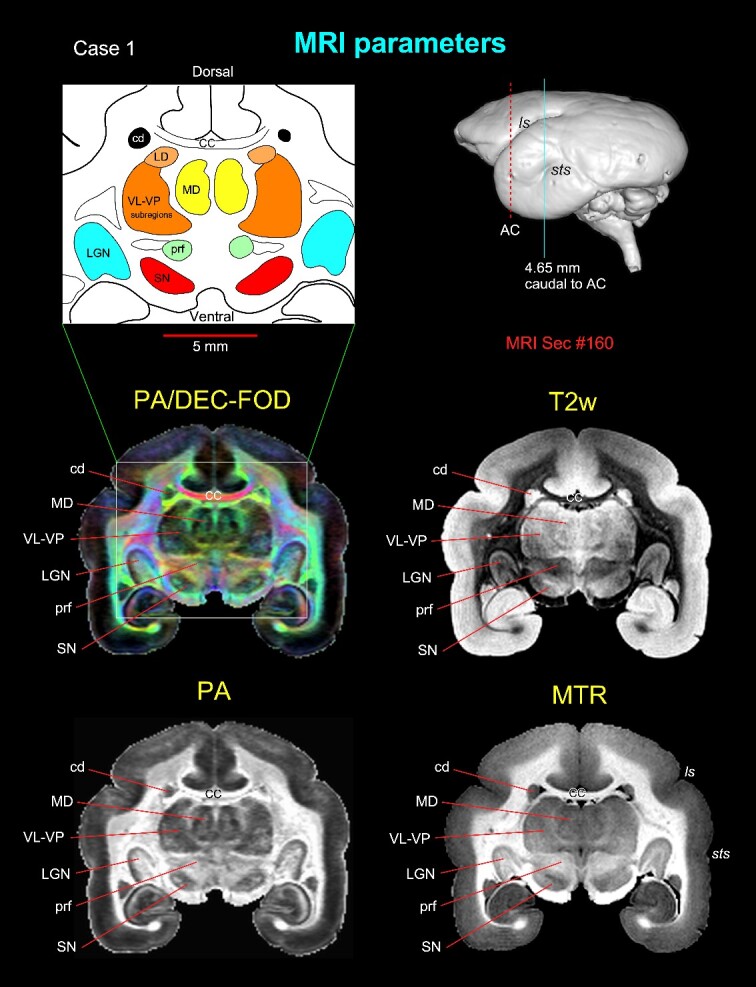
Subcortical regions with different MRI parameters. Matched coronal MR images from 2 of the 8 MAP-MRI parameters, T2-weighted (T2W), and MTR images show selected subcortical regions: thalamic subregions (MD, LD, VL, VP, LGN), basal ganglia subregions (SN, cd), and a prerubral region (prf) anterior to the red nucleus. These areas are also illustrated in the corresponding drawing from the PA/DEC-FOD image on the top left (white box/inset). For mapping and a detailed description of the thalamic and other subcortical regions and other MAP-MRI parameters, see the Results section in Saleem et al. (2023). This MRI slice is located at the level of the rostral temporal cortex and 4.65 mm caudal to the AC, as illustrated by a blue vertical line on the lateral view of the 3D rendered brain image from this case. Note that the contrast between these subcortical areas is distinct in different MRI parameters. Abbreviations: CC, corpus callosum; cd, caudate nucleus; LD, lateral dorsal nucleus; LGN, lateral geniculate nucleus; MD, medial dorsal thalamic nuclei; prf, prerubral field; SN, substantia nigra; subregions of VL, ventral lateral and VP, ventral posterior nuclei. Sulci: ls, lateral sulcus; sts, superior temporal sulcus.
We then converted the delineated two-dimensional subcortical/deep brain regions into a 3D volume. We adapted this new ex vivo 3D volume with 251 delineated regions as “SAM” (Figs 4E–F and 5). This ex vivo atlas was then integrated into the AFNI (Cox 1996; Saad and Reynolds 2012) and SUMA (Surface Mapper (Cox 1996; Saad and Reynolds 2012) software packages with subcortical area labels. To preserve the contiguity of the regions, the subcortical regions were modally smoothed with a simple regularization procedure where each voxel was replaced with the most common voxel label in the immediate neighborhood around each voxel (27 voxels). This procedure was used in our recent macaque D99 atlas version 2.0 (Saleem et al. 2021). The method smooths edges caused by mismatches in 2D drawings applied to a 3D shape. The dataset was then subjected to manual verification and correction of the areal extent and architectonic borders of different subcortical areas, again aided by histology (see above). Additionally, the dataset was tested for “lost clusters” using AFNI’s @ROI_decluster. The “lost clusters” are voxels that are not connected to the main part of the region. Each region is tested to see if it is made of multiple clusters of neighboring voxels. In almost every case, regions are expected to contain only a single clustered set of connected, neighboring voxels. If there are more, then these are commonly small errors in the drawing. While modal smoothing helps to avoid smaller errors like these, it does not remove larger displacements. For this type of error, the differences required manual examination of these disconnected voxels, with replacement to the correct voxel value. In this procedure, each region was clustered to a minimum of half the total voxels for that label. The declustered dataset was compared with the original input volume, and differences were manually corrected and reverified for lost clusters.
Fig. 5.
Symmetric ex vivo SAM atlas and template. (A–C) The “SAM” digital atlas overlaid on the coronal, horizontal, and sagittal ex-vivo T2W MRI templates, respectively. The crosshairs in A–C show the location of the midline thalamic subregion clc (central latocellular nucleus). (D–E) The spatial location of the segmented subcortical regions is shown on the lateral and dorsal views in 3D. The selected subcortical regions in (D–E) are also indicated in (A–C). Abbreviations: CBT, corticobulbar tract; CST, corticospinal tract; mlf, medial longitudinal fasciculus; PAG, periaqueductal gray; POA, preoptic area; SCP, superior cerebellar peduncle. Orientation: D, dorsal; V, ventral; R, rostral; C, caudal; L, lateral.
To create a symmetric template, the left-side brain T2-weighted dataset was mirrored in its x-axis (3dLRflip), and the results were combined to create a symmetric brain. The parcellated atlas was similarly mirrored. Labels were applied to the dataset for AFNI’s whereami functionality to show regions interactively and for command-line region selection. Labels include a short, abbreviated name and a longer descriptive name. Representative center coordinates were assigned based on the left hemisphere regions by an internal center, i.e. the voxel location inside the region closest to the center of mass for that region. Ten empty slices of zero values were added to the inferior, superior, and posterior regions of the dataset for general use as targets in alignment procedures. The datasets were saved in the NIFTI format identified with the NIFTI space code and a brief description, including the atlas version. Labels and dataset template space were saved in the AFNI extension within the NIFTI header.
Registration of the symmetric ex vivo SAM atlas to test subjects
To verify the usefulness and limitations of using an ex vivo template and atlas for in vivo subject marmosets, we registered this new 3D standard symmetric SAM atlas to in vivo T2W MRI volumes of 6 individual (control) marmoset brains of different age groups ranging from 1 to 10 and a multi-subject population-based in vivo T1W template oriented to Ear Bar Zero (EBZ) stereotaxic coordinates (Liu et al. 2021) using the @animal_warper program in AFNI (Jung et al. 2021). The dataset was aligned to the MTR/MAP template using center-shifting, affine, and nonlinear warp transformations. The inverted transformations were combined and applied to the atlas to bring the atlas segmentation to the native space of each marmoset. The default modal smoothing was applied to replace each voxel with the most common neighbor in the immediate 27-voxel neighborhood. For more details, see the Results section and related Fig. 7. The MR scanning methods used to obtain T2W and T1W images of these control animals are described in the following publications (Liu et al. 2021; Hata et al. 2023).
Fig. 7.
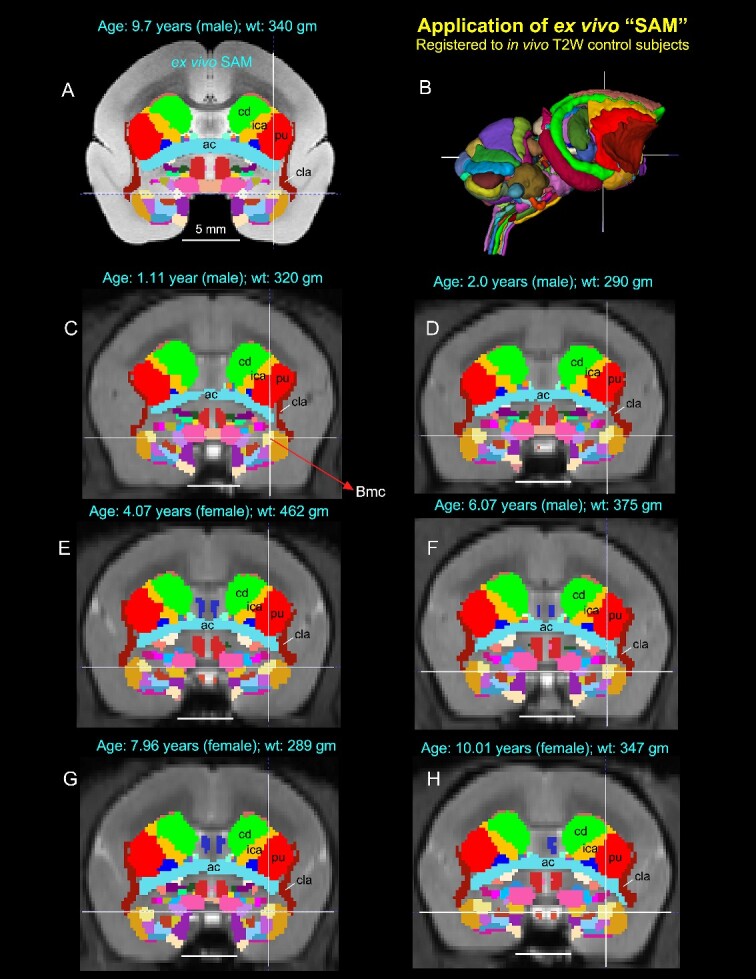
Application of 3D atlas in control subjects. Registration of the SAM digital atlas (B) to various in vivo T2W test subjects of different age groups, genders, and weights using a novel processing pipeline developed within AFNI (see the Method section). (A) Mid-coronal section from the SAM atlas with delineated subcortical regions. (C–H) Coronal slices from 6 control animals, with the SAM atlas registered to the T2W MRI volume of each animal in its native space. None of the registered regions were altered or adjusted in these animals. Note that the corresponding location of the deep brain regions in the control subjects (e.g. ac, anterior commissure; Bmc, basal nucleus of the amygdala, magnocellular division, indicated by cross-hair; cd, caudate; cla, claustrum; ica, anterior limb of the internal capsule; pu, putamen) closely matched with the SAM (A). The MRI volumes of these 6 control subjects were obtained from a publicly shared multimodal brain MRI database covering marmosets with a wide age range (Hata et al. 2023). Scale bars in A–H = 5 mm.
Results
MRI markers
Using high-resolution MRI, we first identified and delineated different subcortical regions for the 3D atlas (Fig. 2). MAP-MRI with different microstructural parameters and other MRI parameters showed different gray and white matter contrasts outside the cerebral cortex. In particular, PA or PA/DEC-FOD (see below), T2W, and MTR (T1-like contrast) images revealed sharp boundaries and high contrast in the deep brain regions, resulting in a clear demarcation of anatomical structures such as the thalamus and other brain regions (e.g. lateral geniculate nucleus (LGN), medial dorsal nucleus of thalamus (MD), prerubral field (prf), and substantia nigra (SN)) (Fig. 2). We have illustrated only a few MRI parameters in Fig. 2, but for other MAP-MRI microstructural parameters derived from marmoset brains, see our previous study (Saleem et al. 2023).
Both PA/DEC-FOD and DEC-FOD refer to the same type of contrast: the propagator anisotropy (PA)-modulated direction encoded color (DEC) (Pajevic and Pierpaoli 1999) and FOD function. Similar to our previous publications (Saleem et al. 2021, 2023), we have changed the wording to DEC-FOD throughout the manuscript for consistency. In each DEC-FOD voxel, the color reflects the dominant fiber orientation, whereas the intensity/brightness is proportional to the propagator anisotropy (PA) in that voxel. Thus, DEC-FOD provides a concise and compelling way to visualize the dominant fiber orientations in regions with the highest anisotropy.
Histological markers
The location, borders, and architectonic features of subcortical gray and white matter regions observed on MRI were confirmed using adjacent and matched histology sections with multiple stains derived from the same marmoset brain specimen (Fig. 1). However, we mostly relied on DEC-FOD MR images to identify and delineate fiber bundles of different sizes and orientations (Figs 3 and 4). The immuno- and histochemical staining used in this study labeled different types of neuronal cells- or both cell bodies and fiber bundles in cortical and subcortical regions (See description in Fig. 1). The SMI-32 antibody recognizes a nonphosphorylated epitope of neurofilament H (Sternberger and Sternberger 1983; Goldstein et al. 1987) and stains a subpopulation of pyramidal neurons and their dendritic processes in the monkey cerebral cortex (Hof and Morrison 1995; Saleem and Logothetis 2012). It is also an important marker for a vulnerable subset of pyramidal neurons in the higher cortical areas visualized in the postmortem brain of Alzheimer’s disease cases (Hof et al. 1990; Hof and Morrison 1990; Thangavel et al. 2009). SMI-32 can detect axonal pathology in brain specimens with traumatic brain injury (TBI) (Johnson et al. 2016). The antibody against ChAT recognizes cholinergic neurons and has been a valuable stain for motor neurons in the monkey and human brainstem (e.g. cranial nerve nuclei), (Horn et al. 2018). AchE is an enzyme that catalyzes the breakdown of acetylcholine and is a valuable marker for delineating different cortical areas (Carmichael and Price 1994) and major subcortical nuclei in the thalamus and brainstem (Jones 1998; Horn et al. 2018). The calcium-binding protein PV is thought to play an important role in intracellular calcium homeostasis. The antibody against PV has been shown to recognize different types of neurons in subcortical regions and a subpopulation of non-pyramidal neurons (GABAergic) in the monkey neocortex (Jones and Hendry 1989; Jones 1998; Saleem et al. 2007). The integrated multimodal approach using multiple histological stains and various MRI parameters enabled detailed noninvasive anatomical mapping and delineation of nuclei and their subregions in deep brain regions.
Delineation of selected subcortical gray and white matter regions for 3D atlas
Using a combined ex vivo MRI and histology (Figs. 1 and 2), we identified and delineated 211 gray matter subregions in deep brain structures, including the basal ganglia, thalamus, hypothalamus, brainstem (midbrain, pons, and medulla), amygdala, bed nucleus of stria terminalis (BST), and basal forebrain (e.g., Figs. 3 and 4). These 211 delineated areas also include architectonically and functionally distinct non-subcortical regions, such as different lobules of the cerebellar cortex and hippocampal formation (HF). In addition, we distinguished and segmented 40 fiber tracts of different orientations and sizes associated with the basal ganglia, thalamus, brainstem, and cerebellum (see Table 1). The examples in Figs 3 and 4A–D illustrate the subcortical gray and white matter regions in MAP-MRI (DEC-FOD) that are segmented with reference to matched histological sections for the 3D atlas. Although we delineated and segmented 251 gray and white matter regions to create a 3D digital template atlas, it is beyond the scope of this study to describe and illustrate all the identified areas in this report. However, we described the detailed mapping and architectonic features of many of these delineated areas in our previous publication (Saleem et al. 2023). We then generated a 3D atlas from these segmented regions, and this newly segmented volume is called ex vivo “SAM,”. Figure 4F shows the lateral view of the SAM with segmented subcortical regions in 3D, superimposed on the rendered brain volume from this case.
Table 1.
Subcortical Atlas of the Marmoset (“SAM”)—Version 1.0
| Atlas label # | Abbreviations | Basal ganglia subregions and associated fiber bundles and Claustrum | |
| 1 | pu | Putamen | Striatum |
| 2 | cd | Caudate nucleus | Striatum |
| 3 | NA | Nucleus accumbens | Ventral striatum |
| 210 | OT | Olfactory tubercle | Ventral striatum |
| 10 | VP | Ventral pallidum | Pallidum |
| 4 | GPe | Globus pallidus external segment | Pallidum |
| 5 | GPi | Globus pallidus internal segment | Pallidum |
| 9 | STN | Subthalamic nucleus | Nigral region |
| 6 | SNpr | Substantia nigra pars reticulata | Nigral region |
| 7 | SNpc | Substantia nigra pars compacta | Nigral region |
| 8 | SNpl | Substantia nigra pars lateralis | Nigral region |
| 70 | SNpm | Substantia nigra pars mixta | Nigral region |
| 215 | lf_al | Lenticular fasciculus and ansa lenticularis | Fiber bundle |
| 213 | H1 | H1 field of Forel | Fiber bundle |
| 212 | H2 | H2 field of Forel | Fiber bundle |
| 211 | mb | Muratoff bundle | Fiber bundle |
| 66 | ac | Anterior commissure | Fiber bundle |
| 474 | cla | Claustrum | other subcortical region |
| Red nucleus | |||
| 526 | prf | Prerubral field | Brainstem/Midbrain |
| 41 | RNpc | Red nucleus, parvicellular division | Brainstem/Midbrain |
| 122 | RNmc | Red nucleus, magnocellular division | Brainstem/Midbrain |
| Thalamic subregions and associated fiber bundles | |||
| 11 | AV | Anterior ventral nucleus | Dorsal thalamus_anterior group |
| 12 | AM | Anterior medial nucleus | Dorsal thalamus_anterior group |
| 128 | AD | Anterior dorsal nucleus | Dorsal thalamus_anterior group |
| 15 | LD | Lateral dorsal nucleus | Dorsal thalamus_anterior group |
| 13 | VApc | Ventral anterior nucleus, parvicellular division | Dorsal thalamus_lateral group |
| 14 | VAmc | Ventral anterior nucleus, magnocellular division | Dorsal thalamus_lateral group |
| 19 | VLc | Ventral lateral caudal nucleus | Dorsal thalamus_lateral group |
| 20 | VLo | Ventral lateral oral nucleus | Dorsal thalamus_lateral group |
| 21 | VPI | Ventral posterior inferior nucleus | Dorsal thalamus_lateral group |
| 22 | VPM | Ventral posterior medial nucleus | Dorsal thalamus_lateral group |
| 23 | VPMpc | Ventral posterior medial nucleus, parvicellular division | Dorsal thalamus_lateral group |
| 26 | LP | Lateral posterior nucleus | Dorsal thalamus_lateral group |
| 27 | VPLc | Ventral posterior lateral caudal nucleus | Dorsal thalamus_lateral group |
| 103 | VLps | Ventral lateral postrema nucleus | Dorsal thalamus_lateral group |
| 127 | pt | Parataenial nucleus | Dorsal thalamus_medial group |
| 38 | Pa | Paraventricular nucleus | Dorsal thalamus_medial group |
| 16 | MDmc | Mediodorsal nucleus, magnocellular division | Dorsal thalamus_medial group |
| 17 | MDpc | Mediodorsal nucleus, parvicellular division | Dorsal thalamus_medial group |
| 18 | MDmf | Mediodorsal nucleus, multiform division | Dorsal thalamus_medial group |
| 31 | MDdc | Mediodorsal nucleus, densocellular division | Dorsal thalamus_medial group |
| 228 | cif | Central inferior nucleus | Dorsal thalamus_medial group |
| 229 | cdc | Central dorsocellular nucleus | Dorsal thalamus_medial group |
| 131 | Re | Reunions nucleus | Dorsal thalamus_medial group |
| 106 | cim | Central intermediate nucleus | Dorsal thalamus_medial group |
| 24 | cnMD | Centromedian nucleus | Dorsal thalamus_intralaminar group |
| 25 | Pf | Parafascicular nucleus | Dorsal thalamus_intralaminar group |
| 104 | csl | Central superior lateral nucleus | Dorsal thalamus_intralaminar group |
| 105 | cl | Central lateral nucleus | Dorsal thalamus_intralaminar group |
| 129 | pcn | Paracentral nucleus | Dorsal thalamus_intralaminar group |
| 130 | clc | Central latocellular nucleus | Dorsal thalamus_intralaminar group |
| 28 | PM | Medial pulvinar | Dorsal thalamus_posterior group |
| 29 | PL | Lateral pulvinar | Dorsal thalamus_posterior group |
| 523 | PIc | Inferior pulvinar, central nucleus | Dorsal thalamus_posterior group |
| 524 | PIm | Inferior pulvinar, medial nucleus | Dorsal thalamus_posterior group |
| 525 | PIp | Inferior pulvinar, posterior nucleus | Dorsal thalamus_posterior group |
| 132 | Pulo | Pulvinar oralis nucleus | Dorsal thalamus_posterior group |
| 135 | ptg | Posterior thalamic group | Dorsal thalamus_posterior group |
| 32 | LGN | Lateral geniculate nucleus | Geniculate region |
| 510 | MGv | Medial geniculate nucleus, ventral division | Geniculate region |
| 511 | MGad | Medial geniculate nucleus, anterodorsal division | Geniculate region |
| 512 | MGm | Medial geniculate nucleus, medial division | Geniculate region |
| 513 | MGz | Medial geniculate nucleus, transition zone | Geniculate region |
| 514 | MGpd | Medial geniculate nucleus, posterodorsal division | Geniculate region |
| 34 | Hl | Lateral habenular nucleus | Epithalamus |
| 35 | Hm | Medial habenular nucleus | Epithalamus |
| 136 | Sg | Suprageniculate nucleus | Other thalamic region |
| 36 | r | Reticular nucleus | Other thalamic region |
| 37 | zic | Zona incerta | Other thalamic region |
| 126 | Sm | Stria medullaris | Fiber bundle |
| 175 | iml | Internal medullary lamina | Fiber bundle |
| Hypothalamic subregions and associated fiber bundles | |||
| 160 | POA | Preoptic area | anterior group |
| 161 | PVN | Paraventricular nucleus | anterior group |
| 162 | SON | Supraoptic nucleus | anterior group |
| 163 | SCN | Suprachiasmatic nucleus | anterior group |
| 164 | DM | Dorsomedial hypothalamic nucleus | Middle (Tuberal and medial) group |
| 165 | VM | Ventromedial hypothalamic nucleus | Middle (Tuberal and medial) group |
| 166 | LT | Lateral hypothalamic nucleus | Middle (Tuberal and medial) group |
| 167 | Arh | Arcuate hypothalamic nucleus | Middle (Tuberal and medial) group |
| 168 | SOR | Supraoptic nucleus, retrochiasmatic part | Middle (Tuberal and medial) group |
| 243 | mtn | Medial tuberal nucleus | Middle (Tuberal and medial) group |
| 193 | TCA | Tuber cinereum area | Middle (Tuberal and medial) group |
| 243 | mtn | Medial tuberal nucleus | Middle (Tuberal and medial) group |
| 170 | PMN | Paramammillary nucleus | Posterior group |
| 39 | MMN | Medial mammillary nucleus | Posterior group |
| 40 | LMN | Lateral mammillary nucleus | Posterior group |
| 191 | SMN | Supramammillary nucleus | Posterior group |
| 67 | OC | Optic chiasm | Fiber bundle |
| 68 | ot | Optic tract | Fiber bundle |
| Brainstem (Midbrain, Pons, and Medulla)/Spinal cord junction and associated fiber bundles | |||
| 42 | 3rd | Third cranial (oculomotor) nuclei | Midbrain |
| 43 | 4th | Fourth cranial (trochlear) nuclei | Midbrain |
| 44 | SC | Superior colliculus | Midbrain |
| 45 | IC | Inferior colliculus | Midbrain |
| 226 | DR | Dorsal raphe | Midbrain |
| 227 | DTg | Dorsal tegmental nucleus | Midbrain |
| 133 | PAG | Periaqueductal gray | Midbrain |
| 138 | inc | Interstitial nucleus of Cajal | Midbrain |
| 139 | nd | Nucleus of Darkschewitsch | Midbrain |
| 140 | IPN | Interpeduncular nucleus | Midbrain |
| 142 | VTA | Ventral tegmental area | Midbrain |
| 159 | PN | Pontine nuclei | Midbrain |
| 177 | MR | Median raphe | Midbrain |
| 180 | vnll | Ventral nucleus of lateral lemniscus | Midbrain |
| 181 | dnll | Dorsal nucleus of lateral lemniscus | Midbrain |
| 268 | APN | Anterior pretectal nucleus | Midbrain |
| 269 | MRF | Midbrain reticular formation | Midbrain |
| 183 | nbic | Nucleus of brachium of inferior colliculus | Midbrain |
| 184 | pbgn | Parabigeminal nucleus | Midbrain |
| 189 | ldtn | Lateral dorsal tegmental nucleus | Midbrain |
| 273 | ltn | Lateral terminal nucleus | Midbrain |
| 274 | atn | Anterior tegmental nucleus | Midbrain |
| 275 | San | Sagulum nucleus | Midbrain/Pons |
| 276 | PRF | Pontine reticular formation | Midbrain/Pons |
| 46 | 5th | Fifth cranial (trigeminal) nuclei | Pons |
| 47 | 6th | Sixth cranial (abducent) nuclei | Pons |
| 48 | 7th | Seventh cranial (facial) nuclei | Pons |
| 52 | soc | Superior olivary complex | Pons |
| 218 | lc | Locus coeruleus | Pons |
| 220 | MC5_nu | Mesencephalic trigeminal nucleus | Pons |
| 221 | PBN | Parabrachial nucleus | Pons |
| 251 | sVN | Superior vestibular nucleus | Pons |
| 252 | pstn | Principal sensory trigeminal nucleus | Pons |
| 254 | RF (ngc) | Reticular formation, nucleus gigantocellularis | Pons |
| 257 | nro | Nucleus raphe obscurus | Pons |
| 258 | nrm | Nucleus raphe magnus | Pons |
| 259 | nrp | Nucleus raphe pallidus | Pons |
| 260 | RF (npo) | Reticular formation, nucleus pontis centralis oralis | Pons |
| 270 | dpgn | Dorsal paragigantocellular nucleus | Pons |
| 271 | lpgn | Lateral paragigantocellular nucleus | Pons |
| 272 | nrip | Nucleus raphe interpositus | Pons |
| 267 | rtn | Reticulo-tegmental nucleus | Pons |
| 528 | ntb | Nucleus of trapezoid body | Pons |
| 50 | stn | Spinal trigeminal nucleus | pons and medulla |
| 248 | mVN | Medial vestibular nucleus | Pons and medulla |
| 249 | lVN | Lateral vestibular nucleus | Pons and medulla |
| 250 | spVN | Spinal vestibular nucleus | Pons and medulla |
| 51 | ion | Inferior olivary nucleus | Medulla |
| 53 | chn | Cochlear nucleus | Medulla |
| 54 | 12th | Twelfth cranial (hypoglossal) nuclei | Medulla |
| 55 | denv_10 | Dorsal motor nucleus of vagus (tenth cranial nuclei) | Medulla |
| 56 | sn | Solitary nucleus | Medulla |
| 57 | np | Nucleus prepositus | Medulla |
| 230 | AN | Ambiguus nucleus | Medulla |
| 58 | gn | Gracile nucleus | Medulla |
| 59 | cn | Cuneate nucleus | Medulla |
| 60 | lcn | Lateral cuneate nucleus | Medulla |
| 232 | lrn | Lateral reticular nucleus | Medulla |
| 255 | irn | Intermediate reticular nucleus | Medulla |
| 256 | prn | Parvicellular reticular nucleus | Medulla |
| 261 | mrn_d | Medullary reticular nucleus, dorsal part | Medulla |
| 262 | mrn_v | Medullary reticular nucleus, ventral part | Medulla |
| 265 | stn (g) | Spinal trigeminal nucleus, gelatinosa layer | Spinal cord |
| 266 | IX-lamina 9 | Lamina 9 of the spinal gray | Spinal cord |
| 278 | CG | Central gray | Spinal cord |
| 134 | bsc | Brachium of superior colliculus | Fiber bundle |
| 144 | mlf | Medial longitudinal fasciculus | Fiber bundle |
| 145 | SCPX | Superior cerebellar peduncle decussation | Fiber bundle |
| 147 | gf | Gracile fasciculus | Fiber bundle |
| 148 | ml | Medial lemniscus | Fiber bundle |
| 149 | tb | Trapezoid body | Fiber bundle |
| 152 | cf | Cuneate fasciculus | Fiber bundle |
| 153 | SCP | Superior cerebellar peduncle | Fiber bundle |
| 154 | pc | Posterior commissure | Fiber bundle |
| 157 | ic_CP_CST_CBT | Internal capsule, cerebral peduncle, corticospinal, or corticobulbar tract | Fiber bundle |
| 182 | bic | Brachium of inferior colliculus | Fiber bundle |
| 185 | ll | Lateral lemniscus | Fiber bundle |
| 190 | MTT | Mammillothalamic tract | Fiber bundle |
| 216 | ICP | Inferior cerebellar peduncle | Fiber bundle |
| 217 | MCP | Middle cerebellar peduncle | Fiber bundle |
| 219 | MC5_t | Mesencephalic trigeminal tract | Fiber bundle |
| 263 | LCST | Lateral corticospinal tract | Fiber bundle |
| 277 | ACST | Anterior corticospinal tract | Fiber bundle |
| 264 | pd | Pyramidal decussation | Fiber bundle |
| 527 | 5n | Motor root of trigeminal nerve | Fiber bundle |
| 529 | 7n | Facial nerve | Fiber bundle |
| 530 | fc | Facial colliculus | Fiber bundle |
| 531 | st | Solitary tract | Fiber bundle |
| 244 | vsct | Ventral spinocerebellar tract | Fiber bundle |
| 245 | cic | Commissure of inferior colliculus | Fiber bundle |
| 246 | csc | Commissure of superior colliculus | Fiber bundle |
| 247 | oct | Olivocerebellar tract | Fiber bundle |
| 253 | 8n | Vestibulocochlear nerve | Fiber bundle |
| 231 | stt | Spinal trigeminal tract_5th | Fiber bundle |
| 69 | fr | Fasciculus retroflexus | Fiber bundle |
| Cerebellum | |||
| 108 | I | Cerebellar lobule I | Lobe |
| 109 | II | Cerebellar lobule II | Lobe |
| 110 | III | Cerebellar lobule III | Lobe |
| 111 | IV | Cerebellar lobule IV | Lobe |
| 112 | V | Cerebellar lobule V | Lobe |
| 113 | VI | Cerebellar lobule VI | Lobe |
| 114 | VII | Cerebellar lobule VII | Lobe |
| 115 | VIII | Cerebellar lobule VIII | Lobe |
| 116 | IX | Cerebellar lobule IX | Lobe |
| 117 | X | Cerebellar lobule X | Lobe |
| 118 | Par_p | Paramedian lobule posterior part | Lobe |
| 119 | Par_a | Paramedian lobule anterior part | Lobe |
| 120 | Cr_II | Crus II of the ansiform lobule | Lobe |
| 121 | Cr_I | Crus I of the ansiform lobule | Lobe |
| 125 | Fl | Flocculus | Lobe |
| 233 | PFl | Paraflocculus | Lobe |
| 222 | Sim_a | Anterior part of the simple lobule | Lobe |
| 223 | Sim_p | Posterior part of the simple lobule | Lobe |
| 61 | DN | Dentate nucleus | Deep cerebellar nuclei |
| 62 | AIN | Anterior interposed nucleus | Deep cerebellar nuclei |
| 63 | PIN | Posterior interposed nucleus | Deep cerebellar nuclei |
| 64 | FN | Fastigial nucleus | Deep cerebellar nuclei |
| Limbic region (Amygdala) | |||
| 237 | L | Lateral nucleus of amygdala | |
| 82 | Bmc | Basal nucleus of amygdala, magnocellular subdivision | |
| 83 | Bi | Basal nucleus of amygdala, intermediate subdivision | |
| 84 | Bpc | Basal nucleus of amygdala, parvicellular subdivision | |
| 85 | ABmc | Accessory basal nucleus of amygdala, magnocellular | |
| 86 | ABpc | Accessory basal nucleus of amygdala, parvicellular | |
| 238 | ABs | Accessory basal nucleus of amygdala, superficial division | |
| 311 | AAA | Anterior amygdaloid area | |
| 87 | COa | Anterior cortical nucleus | |
| 97 | COp | Posterior cortical nucleus | |
| 89 | NLOT | Nucleus of the lateral olfactory tract | |
| 508 | PACo/Pir | Periamygdaloid cortex o/piriform cortex | |
| 509 | PAC2 | Periamygdaloid cortex 2 | |
| 90 | PAC3 | Periamygdaloid cortex 3 | |
| 91 | PACs | Periamygdaloid cortex, sulcal portion | |
| 93 | ME | Medial nucleus of amygdala | |
| 95 | AHA | Amygdalohippocampal area | |
| 96 | PL_prime | Paralaminar nucleus in amygdala | |
| 239 | CEm | Central nucleus of amygdala, medial division | |
| 240 | CEl | Central nucleus of amygdala, lateral division | |
| 241 | CEc | Central nucleus of amygdala, capsular division | |
| Allocortical region (Hippocampal formation) and associated fiber bundle | |||
| 242 | FD | Fascia dentata (granule cell layer) | Dentate Gyrus subregions |
| 71 | CA1 | CA1 subfield of hippocampus | Cornu Ammonis (CA) |
| 72 | CA2 | CA2 subfield of hippocampus | Cornu Ammonis (CA) |
| 73 | CA3 | CA3 subfield of hippocampus | Cornu Ammonis (CA) |
| 74 | CA4 | CA4 subfield of hippocampus | Cornu Ammonis (CA) |
| 76 | preS | Presubiculum | Subicular region |
| 77 | Sub | Subiculum | Subicular region |
| 78 | proS | Prosubiculum | Subicular region |
| 79 | paraS | Parasubiculum | Subicular region |
| 137 | f | Fornix | Fiber bundle |
| Basal forebrain (Cholinergic cell groups) | |||
| 209 | Ch1 | Cholinergic cell group 1 | Basal forebrain |
| 202 | Ch2 | Cholinergic cell group 2 (nucleus of vertical limb of the diagonal band) | Basal forebrain |
| 203 | Ch3 | Cholinergic cell group 3 (nucleus of horizontal limb of the diagonal band) | Basal forebrain |
| 204 | Ch4_al | Cholinergic cell group 4, anterior lateral division (nucleus basalis of Meynert) | Basal forebrain |
| 205 | Ch4_am | Cholinergic cell group 4, anterior medial division (nucleus basalis of Meynert) | Basal forebrain |
| 206 | Ch4_id | Cholinergic cell group 4, intermedio-dorsal division (nucleus basalis of Meynert) | Basal forebrain |
| 207 | Ch4_iv | Cholinergic cell group 4, intermedio-ventral division (nucleus basalis of Meynert) | Basal forebrain |
| 208 | Ch4_p | Cholinergic cell group 4, posterior division | Basal forebrain |
| Bed nucleus of stria terminalis | |||
| 196 | BSTM | Medial bed nucleus of stria terminalis | Bed nucleus of stria terminalis |
| 197 | BSTLcn | Lateral bed nucleus of stria terminalis, central subdivision | Bed nucleus of stria terminalis |
| 198 | BSTP | Posterior bed nucleus of stria terminalis | Bed nucleus of stria terminalis |
| 200 | BSTLc | Lateral bed nucleus of stria terminalis, capsular subdivision | Bed nucleus of stria terminalis |
| 201 | BSTLj | Lateral bed nucleus of stria terminalis, juxtacapsular subdivision | Bed nucleus of stria terminalis |
| Subcortical Atlas of Marmoset (SAM) V1.0 - SUMMARY | |||
| Total number of subcortical areas segmented: 251 | |||
| Segmented gray matter structures: 211 | |||
| Segmented white matter fiber bundles: 40 | |||
Note: The 210 subcortical gray matter structures also include non-subcortical regions such as the hippocampal formation and the cerebellum.
Subcortical areas nomenclature
We did not use the Paxinos atlas (Paxinos et al. 2012) as a primary source reference for our marmoset (SAM) atlas; however, we delineated selected brainstem nuclei in our SAM template with reference to this atlas (see below). For the cytoarchitectonic subdivisions of the thalamus, hypothalamus, amygdala, basal forebrain, BST, and other non-subcortical regions (e.g. HF and cerebellum), we adopted different sources as indicated below.
Thalamus: The thalamic subregions in the marmoset are comparable with those in the macaque monkey, and we adapted the terminology of thalamic nuclei similar to that of Olszewski (Olszewski 1952). The thalamus is divided into the dorsal thalamus, epithalamus, and geniculate regions (Jones 1998). The dorsal thalamus is further divided into anterior, medial, lateral, intralaminar, and posterior groups (Fig. 3B–F) and plays significant roles in memory, emotion, motor, arousal, and other sensorimotor functions (Mitchell et al. 2014; Halassa and Kastner 2017; Pergola et al. 2018). The epithalamus is located in the posterior dorsal part of the diencephalon, and its principal gray matter structure is habenular nuclei, which play a pivotal role in reward processing, aversion, and motivation (Hikosaka et al. 2008; Roman et al. 2020). The geniculate region includes lateral and medial geniculate bodies and is an important relay nucleus in the visual and auditory pathways. We delineated these thalamic subregions in our MRI parameters; however, the MAP-MRI parameter PA (propagator anisotropy) with DEC-FOD is particularly useful for identifying different thalamic nuclei and the surrounding fiber tracts of different orientations in the marmoset, as illustrated in Fig. 3B and C.
Hypothalamus: The hypothalamus can be divided rostrocaudally into the rostral, middle (tuberal), and caudal groups (Rempel-Clower and Barbas 1998; Wells et al. 2020) and has strong connections with the orbital and medial prefrontal cortex (Ongur et al. 1998). The MAP-MRI parameters (PA and PA with DEC-FOD) and matched histological sections with different stains are useful for delineating different subnuclei in the hypothalamus (Fig. 3B).
Brainstem: Brainstem nuclei were identified with reference to the photographic atlas of the human brain (DeArmond et al. 1989) and Duvernoy’s atlas of the human brainstem and cerebellum (Naidich et al. 2009). We also used the marmoset brain atlas (Paxinos et al. 2012) to identify selected nuclei in the brainstem. The spatial location of some brainstem nuclei and fiber bundles in our marmoset MRI is comparable with that in macaques (Saleem et al. 2021). High-resolution MAP-MRI (PA with DEC-FOD) and T2W images are instrumental in delineating gray (cranial nerve nuclei) and white matter subregions in the different rostrocaudal extents of the brainstem (midbrain, pons, and medulla) and cerebellum (Fig. 4). We delineated several sensory and motor nuclei and fiber tracts of different sizes and directions in the brainstem on DEC-FOD images with reference to SMI-32 stained sections (Fig. 4A–D). We also delineated the selected brainstem nuclei in coronal high-resolution T2W images with 100 μm resolution from a different case, confirmed with matched histological sections stained with NeuN, SMI-32, and ChAT (Saleem et al. 2023; their Figs. 13 and 14).
Cerebellum: The unique foliation and compartmental organization of the cerebellar cortex prompted us to identify and segment this non-subcortical region in our high-resolution MR images. Similar to the macaque (Saleem et al. 2021), we identified the spatial location of 10 lobules (I-X), paramedian lobules (Par), simple lobule (Sim), ansiform lobules Crus I and II, flocculus (Fl), and paraflocculus (PFl) (see Table 1) with reference to 11 landmarks (9 fissures and 2 sulci) through the rostrocaudal and mediolateral extent of the cerebellum on the sagittal MR images (Saleem et al. 2023; their Fig. 12A–C). The nomenclature and abbreviations used to identify these lobules and landmarks are according to (Larsell 1953) but with some modifications to include a simplified version of the abbreviations for the fissures and sulci.
Amygdala: The nuclear subdivisions of the amygdaloid complex in the marmoset (Araujo Gois Morais et al. 2019) closely resemble those of macaque monkeys (Price et al. 1987; Amaral and Bassett 1989; Pitkanen and Amaral 1998). The deep nuclei of the amygdala are divided into lateral (L), basal (B), accessory basal (AB), and paralaminar (PL’) nuclei. The lateral nucleus is further divided into 4 subregions (dorsal-Ld, lateral-Ll, medial-Lm, and ventral-Lv), which are defined based on cell size and packing density in Nissl and the intensity of neuropil staining in AchE (Araujo Gois Morais et al. 2019). We identified similar subregions in ChAT- and NeuN = stained sections and matched DEC-FOD and T2W images (Saleem et al. 2023; their Fig. 15).
Hippocampal formation: The distinct architectonic characteristics of the HF (hippocampus proper and subicular complex), prompted us to delineate these allocortical areas in our MAP-MRI parameters with reference to various histological stains. Nine distinct subfields were identified (Rosene and Van-Hoesen 1987; Palomero-Gallagher et al. 2020) and delineated within the HF: Fascia dentata (FD), Cornu ammonis (CA1, CA2, CA3, and CA4), prosubiculum (proS), subiculum proper (Sub), presubiculum (preS), and parasubiculum (paraS), and a transition region at the rostral part of the HF, the hippocampal amygdaloid transition area (HATA).
Basal forebrain: We distinguished different groups of cholinergic neurons of the basal forebrain in the Marmoset with ChAT-stained sections using the nomenclature proposed by Mesulam and colleagues (Mesulam et al. 1984). These are Ch1, Ch2 (nucleus of the vertical limb of the diagonal band), Ch3 (nucleus of the horizontal limb of the diagonal band), and Ch4 (nucleus basalis of Meynert). The Ch4 cell group is further divided into 5 sectors: Ch4-al (anterolateral), Ch4-am (anteromedial), Ch4-id (intermedio-dorsal), Ch4-iv (intermedio-ventral), and Ch4-p (posterior group).
Bed nucleus of stria terminalis (BST): The BST forms part of the extended amygdala and comprises lateral, medial, and posterior divisions (deCampo and Fudge 2013): BSTLcn (lateral bed nucleus of stria terminalis, central subdivision); BSTLc (lateral bed nucleus of stria terminalis, capsular subdivision); BSTLj (lateral bed nucleus of stria terminalis, juxtacapsular subdivision); BSTM (medial bed nucleus of stria terminalis); and BSTP (posterior bed nucleus of stria terminalis). We delineated these subregions in both marmoset (SAM) and macaque (D99) atlases using MAP-MRI and histology with different stains.
Ex vivo SAM template
Figure 5A–C illustrates the new symmetrized SAM digital template atlas with segmented subcortical regions in the horizontal, coronal, and sagittal planes of the sections. We mapped 251 areas, including the subregions of the cerebellar cortex and HF, as described above. Figure 5D–E shows the spatial location of the delineated subcortical regions on the dorsal and lateral views in 3D. This new template atlas is intended for use as a reference standard for marmoset neuroanatomical, functional, and connectional imaging studies involving subcortical targets. Using AFNI’s @animal_warper, the SAM atlas can be automatically registered to the 3D anatomical scans from various marmoset individuals (see Figs 6 and 7) and thus used to specify the areal designation relative to the experimental locations of interest. This digital atlas is now available in the AFNI and SUMA analysis packages to register and apply to the brains of other individual marmoset monkeys to guide research applications for which accurate knowledge of areal boundaries is desired.
Fig. 6.
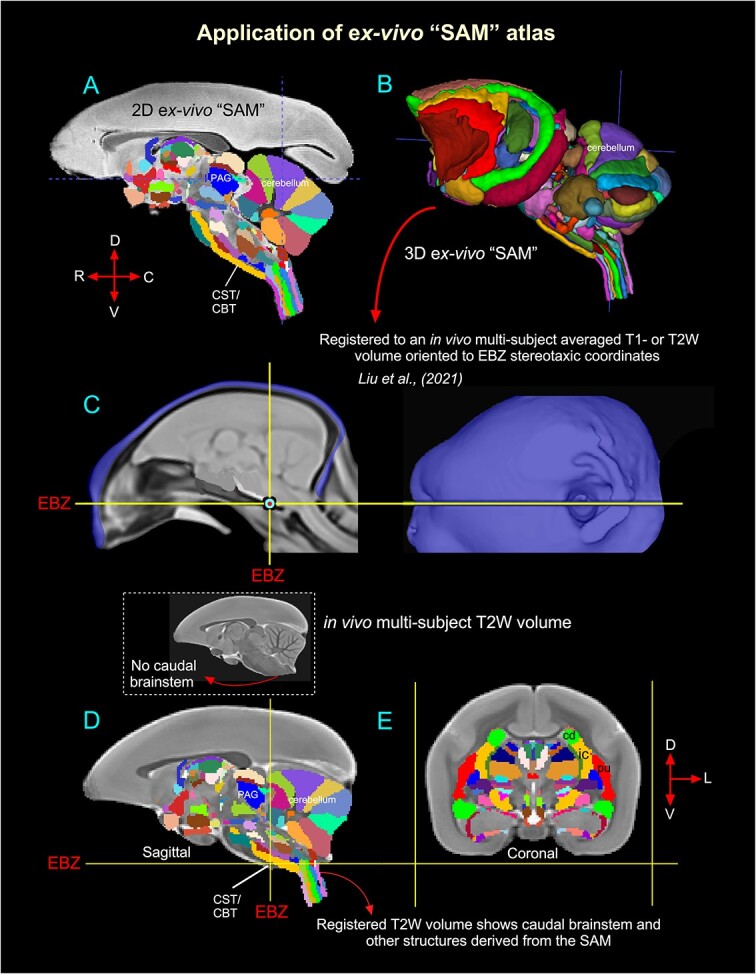
Validation of the ex vivo “SAM” digital template atlas. Two hundred and fifty-one deep brain regions, including the HF and cerebellum, were manually segmented through a series of 150 μm thick MAP-MRI or T2W images (A) using ITK-SNAP and the spatial location of these regions was derived in 3D (B). This new MRI-histology-based segmented volume (called “SAM”) is registered to an in vivo multisubject averaged or population-based T1- or T2W MRI volume (also called MBM template) oriented to EBZ stereotaxic coordinates (Liu et al. 2021) (C). The images in C were obtained and used with permission from the author Cirong Liu (slightly modified for this figure). The illustrations in D–E indicate the registered subcortical areas in the control subject (T2W). None of the registered regions in D and E were altered or adjusted. Note that this control brain specimen (T2W volume) lacks the caudal brainstem (inset with an arrow in D), but after registration with the SAM template, this volume (D) includes the caudal brainstem areas. Abbreviations: CBT, corticobulbar tract; cd, caudate nucleus; CST, corticospinal tract; EBZ, ear bar zero; ic, internal capsule; PAG, periaqueductal gray; pu, putamen. Orientation: D, dorsal; V, ventral; R, rostral; C, caudal; L, lateral.
Application: register identified areas from 3D SAM to a range of test individuals
We estimated and confirmed the atlas-based areal boundaries of subcortical areas by registering this standard ex vivo SAM template with multiple in vivo MRI datasets of different age groups (marmoset control adults). To this end, we developed a novel processing pipeline within AFNI and SUMA to optimally register this SAM atlas to an in vivo T1- or T2W population-based standard template (Liu et al. 2021) or in vivo T2W individual marmoset brain volumes (Hata et al. 2023) (Figs. 6 and 7). This procedure involved a sequence of affine and nonlinear registration steps. An initial affine step gave approximate scaling and rotation to the template. The affinely registered subject brain was gradually warped to the template by progressively smaller nonlinear warps. This procedure resulted in the subject brain data being registered in the SAM template space. By inverting the combination of affine and nonlinear transformations, the atlas segmentation was warped to the original native space of each subject. The results of this pipeline are illustrated in Figs. 6 and 7. In the first example (Fig. 6), the ex vivo SAM was registered to an in vivo T2W multisubject averaged or population-based standard template (also called MBM template) oriented to the EBZ stereotaxic coordinates (Liu et al. 2021). Figure 6D and E shows that the spatial location of transformed segmented areas (e.g. cerebellar lobules, thalamic nuclei) in this T2W averaged volume corresponds well with the delineated areas in the SAM atlas. This aligned dataset (SAM to multisubject MRI template or MBM) is available as a separate download link along with the main download link for the SAM atlas and other MRI templates (see these 2 online links below).
In another example in Fig. 7, the corresponding location of the registered subcortical regions in the control individuals of different age groups, ranging from 1 to 10 yr (e.g. Bmc, magnocellular subdivision of the basal nucleus in the amygdala; cla, claustrum; cd, caudate; pu, putamen; ica, anterior limb of the internal capsule) matched well with the SAM atlas (Fig. 7A–H). While determining the precise matching between the determined areas and the histologically identified regions of all 6 animals is a large project beyond the scope of the present report, these results demonstrate that a straightforward affine and nonlinear warping is sufficient to distinguish and provide atlas-based estimates of areal boundaries in marmoset subjects in vivo.
Atlases and templates are available as volumes and surfaces in standard NIFTI and GIFTI formats. Although this 3D digital atlas can be used in other image registration and analysis software packages, we use the AFNI and SUMA programs with their advanced atlas features for the purposes of demonstration (Cox 1996; Saad and Reynolds 2012). The atlas is integrated into the most recent versions of AFNI and SUMA, enabling straightforward identification of a real identity in any marmoset subject registered to the template and for the individual marmoset subject in its native space by the inverse transformations. The 3D template volume, atlas, and script for atlas registration of in vivo scans are now available for download through the AFNI and SUMA websites at https://afni.nimh.nih.gov/pub/dist/atlases/marmoset/SAM_Marmoset/SAM_marmoset_subcortical_dist.tgz. The AFNI software can install this simply with the @Install_SAM_Marmoset command.
A separate download link is also included for the SAM atlas aligned to the multisubject MRI or MBM dataset (Fig. 6).
https://afni.nimh.nih.gov/pub/dist/atlases/marmoset/SAM_Marmoset/aw_SAM_to_MBM.tgz
Discussion
Despite its essential role as a research model for human brain development and neurological disorders, the marmoset monkey lacked a comprehensive, well-organized MRI-histology-based atlas of subcortical regions. In this study, we generated a 3D digital template atlas of the marmoset from 251 segmented subcortical regions (called “SAM”) using high-resolution MAP-MRI, T2W, and MTR images, which were combined and correlated with the histological sections of the same brain specimen. Our results demonstrate that, at a high spatial resolution, the combined use of MRI parameters and matched histology sections with 5 different stains enabled detailed noninvasive segmentation of gray and white matter regions in deep brain structures. This integrated multimodal approach yields a more objective and reproducible delineation of nuclei and their boundaries in the deep brain structures, which include the basal ganglia, thalamus, hypothalamus, limbic region (amygdala), basal forebrain, and rostrocaudal extent of the brainstem (midbrain, pons, and medulla). Many of these deep brain targets and their subregions are less prominent or indistinguishable from neighboring structures with conventional T1W or T2W MRI volumes (Saleem et al. 2023). This new atlas is intended for use as a reference standard template for neuroanatomical, functional (fMRI), clinical, and connectional imaging studies involving subcortical targets in marmoset monkeys. We also estimated and confirmed the atlas-based areal boundaries of subcortical areas by registering this ex vivo atlas template to in vivo T1- or T2W MRI datasets of marmoset control adults (single and multisubject population-based volumes), using a novel pipeline developed within AFNI. In the following sections, we compare our new marmoset digital atlas, “SAM,” with other available atlases in the field and highlight some advantages of the present offering.
Standard SAM versus other marmoset MRI- and histology-based atlases
The present marmoset digital template atlas of subcortical regions, derived from MRI and histology, is one of the few digital atlases created in recent years. In one study, Liu and colleagues (Liu et al. 2018) constructed a 3D digital atlas of the marmoset brain based on MR image contrasts observed in ex vivo MTR, T2W, and diffusion MR images. This study manually delineated 54 cortical but only 16 subcortical areas in their digital atlas. It also lacks the parcellation of brainstem structures (midbrain, pons, and medulla) and nuclei within the major subcortical structures such as the thalamus and amygdala (for example). No histological information is available from this study. Although this MRI-based atlas is helpful for some applications, delineating the cytoarchitectonic areas based on MRI contrasts alone, without corresponding and matched histological information from the same brain specimen to serve as a control, may produce inaccurate boundaries that include additional gray and white matter regions, leading to biases in ROI size and volume estimation. As indicated in our previous macaque (Saleem et al. 2021) and marmoset (Saleem et al. 2023) studies, multimodal MRI parameters acquired with high-spatial resolution (100–200 μm), aided by histology derived from the same brain specimen, are key to delineating nuclei and their subregions for the construction of 3D digital template atlases. In another study, Majka and colleagues (Majka et al. 2020, 2021) created a Nencki–Monash template, a probabilistic atlas based on the morphological average of 20 young marmoset brains obtained by 3D reconstructions generated from Nissl-stained serial sections. It provided a cytoarchitectonic parcellation of cortical areas but no subcortical or deep brain regions.
In a different study, Hashikawa and colleagues (Hashikawa et al. 2015) reconstructed a series of Nissl-stained axial slices into a 3D brain model with cortical and subcortical parcellations using a volume-rendering method. They also reproduced virtual low-resolution parasagittal and coronal slices from this axially generated 3D volume. The introduction of the histology-based template atlases and space was an important step forward and offered a good estimate of areal boundaries and virtual brain structures delineated by their histological features in 3D space. While useful for some applications, this approach does not attempt to preserve the native geometry of the brain because of the distortion and shrinkage of histological sections during section cutting, staining, and mounting. Thus, the spatial accuracy of the resulting volumetric or surface atlas remains questionable. Chemoarchitectonic characterization and parcellation are also necessary for delineating several brain structures in cortical and subcortical regions (Saleem et al. 2021, 2023), but these features are lacking in these studies.
The present digital atlas avoids geometrical transformations of the cytoarchitectonic information from histological sections to match the layout of the animal’s brain. Such transformations can introduce registration errors that are difficult to correct. The most important unique feature of our subcortical atlas (SAM) is the strict adherence to an MRI scan with adjacent and matched histology sections with multiple histo- and immunohistochemical staining from the same brain. As a result, the alignment accuracy between areal boundaries and gross anatomical features is optimized for identifying ROIs in this study (Figs. 3 and 4 in this study; 4–9 in Saleem et al. 2023).
Generalization and validation of 3D atlas
The ex vivo SAM volume is registered to multiple in vivo 3D templates of different age groups using widely available whole-brain MRI registration tools. When applied to the 3D volume, the transformation derived from this warping allows for the labeling of subcortical targets in the brains of individual animals as accurately as possible. It also integrates the information directly with the anatomical and functional imaging results in surface modes. Compiling any brain atlas, which includes the assignment of boundaries and names to individual areas, is an inherently imperfect endeavor whose main goal is to provide a common anatomical framework for a range of research projects and data. In the present case, the innovation rests on the creation of a 3D digital marmoset atlas whose anatomical borders were, from the outset, created on the basis of MR-registered histological sections. This digital atlas (SAM) is based on the precise histological borders from one particular monkey, which, because of the initial registration to the MRI from the same animal, can be represented on the brain of any experimental animal via an alignment procedure such as the one used in this study.
A complete validation of this 3D atlas, such as estimating the architectonic boundaries between different areas for a population of marmoset brains, is beyond the scope of the present study. Nonetheless, our multipronged analysis supports the validity of the SAM template by estimating the architectonic boundaries between different subcortical/deep brain areas for a population of in vivo T1- or T2W marmoset brains. In this analysis, we revealed that the MRI registration procedure using SAM (Figs. 6 and 7) could be smoothly applied to test subjects of different genders, age groups (1–10 yr old), and sizes (290–462 g). Thus, it is possible to estimate the histological boundaries of subcortical areas in any marmoset monkey (Fig. 7). Validation of brain regions in a given subject is helpful for neurosurgical navigation of electrodes or implantable devices to a potential target for DBS in NHP models of psychiatric or neurological disorders (e.g. Parkinson’s disease (Min et al. 2016; Vitek and Johnson 2019). It is also useful for localizing labeled neurons and terminals after anatomical tracer injections, fMRI activation regions (Baker et al. 2006; Logothetis et al. 2012; Turchi et al. 2018; Murris et al. 2020), and mapping the trajectories of subcortical development from young to adult marmoset monkeys (Seki et al. 2017; Sawiak et al. 2018).
MAP-MRI-based atlases in humans (future directions)
In this study, we delineated and generated a 3D SAM monkey using high-resolution MAP-MRI and other MRI parameters and matched histological sections with multiple stains derived from the same brain specimen. We further illustrated how our atlas can be used to locate small subcortical structures after registration to T1- or T2W MRI volumes of control subjects acquired in vivo. These results indicate the utility of a high-resolution atlas in studies of marmoset monkey disease models and highlight the potential of high-resolution MAP-MRI in delineating small subcortical structures based on differences in microstructural properties. The spatial resolution of MAP-MRI data acquired on clinical scanners is significantly lower, ~1 to 2 mm, than that used here. Nevertheless, promising advances in gradient coil design (McNab et al. 2013), radiofrequency (RF) coil engineering (Keil et al. 2013; Truong et al. 2014), dMRI pulse sequence design (Avram et al. 2014b), and spatial encoding (Feinberg et al. 2010; Setsompop et al. 2018) are expected to significantly improve spatial resolution and signal-to-noise ratio to allow submillimeter clinical MAP-MRI scans in the near future (Huang et al. 2021). Concurrently, new clinically feasible diffusion encoding strategies (Avram et al. 2010, 2019, 2021) and analyses (Avram et al. 2022a; Magdoom et al. 2023) are being developed to characterize and delineate specific microscopic tissue water pools without the need to increase spatial resolutions. Taken together, these advances will enable the construction of high-resolution cortical and subcortical maps and atlas templates of the human brain that will improve the localization of neurosurgical navigation, fMRI responses, and high-precision placement of recording and stimulating electrodes in patients with Parkinson's disease, epilepsy, mild TBI, and other diseases.
Summary and conclusion
We have created a comprehensive MRI template and corresponding digital atlas of subcortical regions in the marmoset brain based on a large set of histological and very high-resolution structural MRI and MAP-MRI data for a single marmoset. The atlas provides a usable standard for region definition, while the template provides a standard reference and space. This standard space allows for marmoset research to be reported on a common basis across research sites and across marmoset monkeys. As used in human studies with the Montreal Neurological Institute or Talairach space, this target template space provides a platform for voxelwise group analysis. In addition, the atlas allows for automated analysis against a set of standard region locations, either in the template space or in the native space of the individual subjects. The current atlas, template MRI datasets, surfaces, and user scripts for aligning individual subjects to this template are publicly available at the following link: https://afni.nimh.nih.gov/pub/dist/atlases/marmoset/SAM_Marmoset/SAM_marmoset_subcortical_dist.tgz.
Acknowledgments
We thank James Pickel, the Transgenic core facility at NIMH, for providing a perfusion-fixed marmoset monkey brain for our experiments; Cecil Chern-Chyi Yen for helping with the initial setup for the MRI scanning; and Michal Komlosh for preparing the specimen for MRI. Finally, we thank the Microscope Imaging Core (MIC) at NICHD for helping with the high-resolution imaging of histology sections. All histological processing of the brain tissue was done by Dr Du and his team at FD-NeuroTechnologies in Columbia, Maryland.
Contributor Information
Kadharbatcha S Saleem, Section on Quantitative Imaging and Tissue Sciences (SQITS), Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), National Institute of Health (NIH), 13, South Drive, Bethesda, MD 20892, United States; Military Traumatic Brain Injury Initiative (MTBI2), Henry M. Jackson Foundation for the Advancement of Military Medicine, 6720A Rockledge Drive, Bethesda, MD 20817, United States.
Alexandru V Avram, Section on Quantitative Imaging and Tissue Sciences (SQITS), Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), National Institute of Health (NIH), 13, South Drive, Bethesda, MD 20892, United States.
Daniel Glen, Scientific and Statistical Computing Core, National Institute of Mental Health (NIMH), NIH, 10 Center Drive, Bethesda, MD 20817, United States.
Vincent Schram, Microscopy and Imaging Core (MIC), Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), NIH, 35 Convent Drive, Bethesda, MD 20892, United States.
Peter J Basser, Section on Quantitative Imaging and Tissue Sciences (SQITS), Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), National Institute of Health (NIH), 13, South Drive, Bethesda, MD 20892, United States.
Author contributions
Kadharbatcha S. Saleem: Corresponding author, designed the study, coordinated the project, prepared the specimen for MRI and histology; mapped, segmented, and verified all the anatomical regions of interest with reference to MRI and histology; generated a new subcortical 3D atlas template/Table, made illustrations; and wrote, edited, and streamlined the manuscript (Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Supervision, Validation, Visualization, Writing—original draft, Writing—review & editing).
Alexandru V. Avram: Conducted all MRI experiments, including MAP-MRI scans, processed and analyzed all MRI data, helped with atlas data registration, and wrote and edited the manuscript (Data curation, Methodology, Writing-review & editing).
Daniel Glen: Integrated the atlas dataset into the AFNI and SUMA software packages, wrote the code for atlas region regularization, helped with atlas data registration, and edited the manuscript (Methodology, Software, Visualization, Writing-review & editing).
Vincent Schram: Obtained high-resolution images of all histology-stained sections in the microscope imaging center (MIC) at NICHD and commented on the manuscript (Data curation).
Peter J. Basser: Helped design the study, edited the manuscript, and provided research resources (Conceptualization, Funding acquisition, Resources, Writing-review & editing).
Funding
This work was supported by the CNRM Neuroradiology/Neuropathology Correlation/Integration Core, 309698-4.01-65310 (CNRM-89-9921); Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development; the Intramural Research Program of the National Institute of Neurological Disorders and Stroke; “Connectome 1.0: Developing the next generation human MRI scanner for bridging studies of the micro-, meso -and macro-connectome”, NIH BRAIN Initiative 1U01EB026996-01.
Conflict of interest statement: The authors have no conflicts of interest to disclose. The views, information or content, and conclusions presented do not necessarily represent the official position or policy of, nor should any official endorsement be inferred on the part of the Uniformed Services University, the Department of Defense, the US Government, or the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc.
References
- Abosch A, Yacoub E, Ugurbil K, Harel N. An assessment of current brain targets for deep brain stimulation surgery with susceptibility-weighted imaging at 7 tesla. Neurosurgery. 2010:67:1745–1756 discussion 1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggleton JP, O'Mara SM. The anterior thalamic nuclei: core components of a tripartite episodic memory system. Nat Rev Neurosci. 2022:23(8):505–516. 10.1038/s41583-022-00591-8. [DOI] [PubMed] [Google Scholar]
- Amaral DG, Bassett JL. Cholinergic innervation of the monkey amygdala: an immunohistochemical analysis with antisera to choline acetyltransferase. J Comp Neurol. 1989:281:337–361. [DOI] [PubMed] [Google Scholar]
- Araujo Gois Morais PL, Garcia-Amado M, Lima RRM, Cordoba-Claros A, Souza Cavalcante J, Clasca F, NascimentoES Jr. Cyto- and myelo-architecture of the amygdaloid complex of the common marmoset monkey (Callithrix jacchus). Front Neuroanat. 2019:13:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram AV, Guidon A, Song AW. Myelin water weighted diffusion tensor imaging. NeuroImage. 2010:53:132–138. [DOI] [PubMed] [Google Scholar]
- Avram AV, Barnett AS, Basser PJ. The variation of MAP-MRI derived parameters along white matter fiber pathways in the human brain. International Society for Magnetic Resonance in Medicine (ISMRM); 2014a. p. 2587 [Google Scholar]
- Avram AV, Guidon A, Truong T-K, Liu C, Song AW. Dynamic and inherent B0 correction for DTI using stimulated echo spiral imaging. Magn Reson Med. 2014b:71:1044–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram AV, Sarlls JE, Barnett AS, Ozarslan E, Thomas C, Irfanoglu MO, Hutchinson E, Pierpaoli C, Basser PJ. Clinical feasibility of using mean apparent propagator (MAP) MRI to characterize brain tissue microstructure. NeuroImage. 2016:127:422–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram AV, Hutchinson E, Basser PJ. Higher-order statistics of 3D spin displacement probability distributions measured with MAP MRI. International Society for Magnetic Resonance in Medicine (ISMRM); 2017. p. 3367 [Google Scholar]
- Avram AV, Bernstein AS, Irfanoglu MO, Simmons A, Cota M, Gai N, Jikaria A, Moses A, Turtzo C, Latour L, et al. Anatomical atlas of 3D MAP MRI-derived 3D diffusion propagators and microstructural parameters. International Society for Magnetic Resonance in Medicine (ISMRM); 2018a. p. 1577 [Google Scholar]
- Avram AV, Sarlls JE, Hutchinson E, Basser PJ. Efficient experimental designs for isotropic generalized diffusion tensor MRI (IGDTI). Magn Reson Med. 2018b:79:180–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram AV, Sarlls JE, Basser PJ. Measuring non-parametric distributions of intravoxel mean diffusivities using a clinical MRI scanner. NeuroImage. 2019:185:255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram AV, Sarlls JE, Basser PJ. Whole-brain imaging of subvoxel T1-diffusion correlation spectra in human subjects. Front Neurosci. 2021:15:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram AV, Saleem KS, Basser PJ. COnstrained Reference frame diffusion TEnsor Correlation Spectroscopic (CORTECS) MRI: a practical framework for high-resolution diffusion tensor distribution imaging. Front Neurosci. 2022a:16:1054509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram AV, Saleem KS, Komlosh ME, Yen CC, Ye FQ, Basser PJ. High-resolution cortical MAP-MRI reveals areal borders and laminar substructures observed with histological staining. NeuroImage. 2022b:264:119653. [DOI] [PubMed] [Google Scholar]
- Baker JT, Patel GH, Corbetta M, Snyder LH. Distribution of activity across the monkey cerebral cortical surface, thalamus and midbrain during rapid, visually guided saccades. Cereb Cortex. 2006:16:447–459. [DOI] [PubMed] [Google Scholar]
- Burman KJ, Rosa MG. Architectural subdivisions of medial and orbital frontal cortices in the marmoset monkey (Callithrix jacchus). J Comp Neurol. 2009:514(1):11–29. 10.1002/cne.21976. [DOI] [PubMed] [Google Scholar]
- Carmichael ST, Price JL. Architectonic subdivision of the orbital and medial prefrontal cortex in the macaque monkey. J Comp Neurol. 1994:346:366–402. [DOI] [PubMed] [Google Scholar]
- Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996:29:162–173. [DOI] [PubMed] [Google Scholar]
- DeArmond SJ, Fusco MM, Dewey MM. Structure of the human brain. A photographic atlas. Oxford/New York: Oxford University Press; 1989. [Google Scholar]
- deCampo DM, Fudge JL. Amygdala projections to the lateral bed nucleus of the stria terminalis in the macaque: comparison with ventral striatal afferents. J Comp Neurol. 2013:521(14):3191–3216. 10.1002/cne.23340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deistung A, Schafer A, Schweser F, Biedermann U, Gullmar D, Trampel R, Turner R, Reichenbach JR. High-resolution MR imaging of the human brainstem in vivo at 7 tesla. Front Hum Neurosci. 2013a:7:710. 10.3389/fnhum.2013.00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deistung A, Schafer A, Schweser F, Biedermann U, Turner R, Reichenbach JR. Toward in vivo histology: a comparison of quantitative susceptibility mapping (QSM) with magnitude-, phase-, and R2*-imaging at ultra-high magnetic field strength. NeuroImage. 2013b:65:299–314. [DOI] [PubMed] [Google Scholar]
- Ewert S, Plettig P, Li N, Chakravarty MM, Collins DL, Herrington TM, Kuhn AA, Horn A. Toward defining deep brain stimulation targets in MNI space: a subcortical atlas based on multimodal MRI, histology and structural connectivity. NeuroImage. 2018:170:271–282. [DOI] [PubMed] [Google Scholar]
- Feinberg DA, Moeller S, Smith SM, Auerbach E, Ramanna S, Glasser MF, Miller KL, Ugurbil K, Yacoub E. Multiplexed echo planar imaging for sub-second whole brain FMRI and fast diffusion imaging. PLoS One. 2010:5(12):e15710, 1–11. 10.1371/journal.pone.0015710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrazzoli D, Ortelli P, Madeo G, Giladi N, Petzinger GM, Frazzitta G. Basal ganglia and beyond: the interplay between motor and cognitive aspects in Parkinson's disease rehabilitation. Neurosci Biobehav Rev. 2018:90:294–308. [DOI] [PubMed] [Google Scholar]
- Fukushima M, Ichinohe N, Okano H. Neuroanatomy of the marmoset. Vol. 1. London: Academic Press; 2019. pp. 43–62 [Google Scholar]
- Goldstein ME, Sternberger LA, Sternberger NH. Varying degrees of phosphorylation determine microheterogeneity of the heavy neurofilament polypeptide (Nf-H). J Neuroimmunol. 1987:14:135–148. [DOI] [PubMed] [Google Scholar]
- Halassa MM, Kastner S. Thalamic functions in distributed cognitive control. Nat Neurosci. 2017:20:1669–1679. [DOI] [PubMed] [Google Scholar]
- Hardman CD, Ashwell KWS. Stereotaxic and chemoarchitectural atlas of the brain of the common marmoset (Callithrix jacchus). New York: CRC Press; 2012. [Google Scholar]
- Hashikawa T, Nakatomi R, Iriki A. Current models of the marmoset brain. Neurosci Res. 2015:93:116–127. [DOI] [PubMed] [Google Scholar]
- Hata J, Nakae K, Tsukada H, Woodward A, Haga Y, Iida M, Uematsu A, Seki F, Ichinohe N, Gong R, et al. Multi-modal brain magnetic resonance imaging database covering marmosets with a wide age range. Sci Data. 2023:10:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikosaka O, Sesack SR, Lecourtier L, Shepard PD. Habenula: crossroad between the basal ganglia and the limbic system. J Neurosci. 2008:28:11825–11829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch MJ, Bruno MT, Faustin A, Cruz N, Crandall L, Wisniewski T, Devinsky O, Shepherd TM. 3T MRI whole-brain microscopy discrimination of subcortical anatomy, part 1: brain stem. AJNR Am J Neuroradiol. 2019a:40:401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch MJ, Bruno MT, Faustin A, Cruz N, Mogilner AY, Crandall L, Wisniewski T, Devinsky O, Shepherd TM. 3T MRI whole-brain microscopy discrimination of subcortical anatomy, part 2: basal forebrain. AJNR Am J Neuroradiol. 2019b:40:1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof PR, Morrison JH. Quantitative analysis of a vulnerable subset of pyramidal neurons in Alzheimer's disease: II. Primary and secondary visual cortex. J Comp Neurol. 1990:301:55–64. [DOI] [PubMed] [Google Scholar]
- Hof PR, Morrison JH. Neurofilament protein defines regional patterns of cortical organization in the macaque monkey visual system: a quantitative immunohistochemical analysis. J Comp Neurol. 1995:352:161–186. [DOI] [PubMed] [Google Scholar]
- Hof PR, Cox K, Morrison JH. Quantitative analysis of a vulnerable subset of pyramidal neurons in Alzheimer's disease: I. Superior frontal and inferior temporal cortex. J Comp Neurol. 1990:301(1):44–54. 10.1002/cne.903010105. [DOI] [PubMed] [Google Scholar]
- Horn AKE, Horng A, Buresch N, Messoudi A, Hartig W. Identification of functional cell groups in the abducens nucleus of monkey and human by perineuronal nets and choline acetyltransferase immunolabeling. Front Neuroanat. 2018:12:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SY, Witzel T, Keil B, Scholz A, Davids M, Dietz P, Rummert E, Ramb R, Kirsch JE, Yendiki A, et al. Connectome 2.0: developing the next-generation ultra-high gradient strength human MRI scanner for bridging studies of the micro-, meso- and macro-connectome. NeuroImage. 2021:243:118530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson EB, Schwerin SC, Avram AV, Juliano SL, Pierpaoli C. Diffusion MRI and the detection of alterations following traumatic brain injury. J Neurosci Res. 2018:96:612–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iriki I, Okano HJ, Sasaki E, Okano H. 2018. The 3-dimensional atlas of the marmoset brain-reconstructible in stereotaxic coordinates. Japan: Springer, 10.1007/978-4-431-56612-0. [DOI] [Google Scholar]
- Johnson VE, Stewart W, Weber MT, Cullen DK, Siman R, Smith DH. SNTF immunostaining reveals previously undetected axonal pathology in traumatic brain injury. Acta Neuropathol. 2016:131:115–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EG. The thalamus of primates. In: Handbook of chemical neuroanatomy: the primate nervous system. Part II. New York: Elsevier; 1998. [Google Scholar]
- Jones EG, Hendry SH. Differential calcium binding protein immunoreactivity distinguishes classes of relay neurons in monkey thalamic nuclei. Eur J Neurosci. 1989:1(3):222–246. 10.1111/j.1460-9568.1989.tb00791.x. [DOI] [PubMed] [Google Scholar]
- Jung B, Taylor PA, Seidlitz J, Sponheim C, Perkins P, Ungerleider LG, Glen D, Messinger A. A comprehensive macaque fMRI pipeline and hierarchical atlas. NeuroImage. 2021:235:117997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas JH. Comparative functional anatomy of marmoset brains. ILAR J. 2020:61:260–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil B, Blau JN, Biber S, Hoecht P, Tountcheva V, Setsompop K, Triantafyllou C, Wald LL. A 64-channel 3T array coil for accelerated brain MRI. Magn Reson Med. 2013:70:248–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuken MC, Bazin PL, Crown L, Hootsmans J, Laufer A, Muller-Axt C, Sier R, van der PuttenEJ, Schafer A, Turner R, et al. Quantifying inter-individual anatomical variability in the subcortex using 7 T structural MRI. NeuroImage. 2014:94:40–46. [DOI] [PubMed] [Google Scholar]
- Koay CG, Özarslan E, Johnson KM, Meyerand ME. Sparse and optimal acquisition design for diffusion MRI and beyond. Med Phys. 2012:39:2499–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciego JL, Luquin N, Obeso JA. Functional neuroanatomy of the basal ganglia. Cold Spring Harb Perspect Med. 2012:2:a009621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsell O. The cerebellum of the cat and the monkey. J Comp Neurol. 1953:99:135–199. [DOI] [PubMed] [Google Scholar]
- Lenglet C, Abosch A, Yacoub E, De Martino F, Sapiro G, Harel N. Comprehensive in vivo mapping of the human basal ganglia and thalamic connectome in individuals using 7T MRI. PLoS One. 2012:7:e29153, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Ye FQ, Yen CC, Newman JD, Glen D, Leopold DA, Silva AC. A digital 3D atlas of the marmoset brain based on multi-modal MRI. NeuroImage. 2018:169:106–116. 10.1016/j.neuroimage.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Yen CC, Szczupak D, Tian X, Glen D, Silva AC. Marmoset brain mapping V3: population multi-modal standard volumetric and surface-based templates. NeuroImage. 2021:226:117620. 10.1016/j.neuroimage.2020.117620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logothetis NK, Eschenko O, Murayama Y, Augath M, Steudel T, Evrard HC, Besserve M, Oeltermann A. Hippocampal-cortical interaction during periods of subcortical silence. Nature. 2012:491:547–553. [DOI] [PubMed] [Google Scholar]
- Magdoom KN, Avram AV, Sarlls JE, Dario G, Basser PJ. A novel framework for in-vivo diffusion tensor distribution MRI of the human brain. NeuroImage. 2023:271:120003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka P, Bai S, Bakola S, Bednarek S, Chan JM, Jermakow N, Passarelli L, Reser DH, Theodoni P, Worthy KH, et al. Open access resource for cellular-resolution analyses of corticocortical connectivity in the marmoset monkey. Nat Commun. 2020:11:1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka P, Bednarek S, Chan JM, Jermakow N, Liu C, Saworska G, Worthy KH, Silva AC, Wojcik DK, Rosa MGP. Histology-based average template of the marmoset cortex with probabilistic localization of cytoarchitectural areas. NeuroImage. 2021:226:117625. [DOI] [PubMed] [Google Scholar]
- McNab JA, Edlow BL, Witzel T, Huang SY, Bhat H, Heberlein K, Feiweier T, Liu K, Keil B, Cohen-Adad J, et al. The Human Connectome Project and beyond: initial applications of 300 mT/m gradients. NeuroImage. 2013:80:234–245. 10.1016/j.neuroimage.2013.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesulam MM, Mufson EJ, Levey AI, Wainer BH. Atlas of cholinergic neurons in the forebrain and upper brainstem of the macaque based on monoclonal choline acetyltransferase immunohistochemistry and acetylcholinesterase histochemistry. Neuroscience. 1984:12:669–686. [DOI] [PubMed] [Google Scholar]
- Min HK, Ross EK, Jo HJ, Cho S, Settell ML, Jeong JH, Duffy PS, Chang SY, Bennet KE, Blaha CD, et al. Dopamine release in the nonhuman primate caudate and putamen depends upon site of stimulation in the subthalamic nucleus. J Neurosci. 2016:36:6022–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AS, Sherman SM, Sommer MA, Mair RG, Vertes RP, Chudasama Y. Advances in understanding mechanisms of thalamic relays in cognition and behavior. J Neurosci. 2014:34:15340–15346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murris SR, Arsenault JT, Vanduffel W. Frequency- and state-dependent network effects of electrical stimulation targeting the ventral tegmental area in macaques. Cereb Cortex. 2020:30:4281–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidich TP, Duvernoy HM, Delman BN, Sorensen AG, Kollias SS, Haacke EM. Duvernoy’s atlas of the human brain stem and cerebellum. High-field MRI: surface anatomy, internal structure, vascularization and 3D sectional anatomy. New York: Springer-Wien; 2009. [Google Scholar]
- Newman JD, Kenkel WM, Aronoff EC, Bock NA, Zametkin MR, Silva AC. A combined histological and MRI brain atlas of the common marmoset monkey, Callithrix jacchus. Brain Res Rev. 2009:62(1):1–18. 10.1016/j.brainresrev.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano H, Sasaki E, Yamamori T, Iriki A, Shimogori T, Yamaguchi Y, Kasai K, Miyawaki A. Brain/MINDS: a Japanese National Brain Project for marmoset neuroscience. Neuron. 2016:92:582–590. [DOI] [PubMed] [Google Scholar]
- Olszewski J. The thalamus of the Macaca mulatta: an atlas for use with the stereotaxic instrument. New York, Basel: S. Karger; 1952. [Google Scholar]
- Ongur D, An X, Price JL. Prefrontal cortical projections to the hypothalamus in macaque monkeys. J Comp Neurol. 1998:401(4):480–505. . [DOI] [PubMed] [Google Scholar]
- Ozarslan E, Koay CG, Shepherd TM, Komlosh ME, Irfanoglu MO, Pierpaoli C, Basser PJ. Mean apparent propagator (MAP) MRI: a novel diffusion imaging method for mapping tissue microstructure. NeuroImage. 2013:78:16–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajevic S, Pierpaoli C. Color schemes to represent the orientation of anisotropic tissues from diffusion tensor data: application to white matter fiber tract mapping in the human brain. Magn Reson Med. 1999:42:526–540. [PubMed] [Google Scholar]
- Palazzi X, Bordier N. The marmoset brain in stereotaxic coordinates. New York: Springer Science+Business Media LLC; 2008. [Google Scholar]
- Palomero-Gallagher N, Kedo O, Mohlberg H, Zilles K, Amunts K. Multimodal mapping and analysis of the cyto- and receptorarchitecture of the human hippocampus. Brain Struct Funct. 2020:225:881–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli WM, Nili AN, Tyszka JM. A high-resolution probabilistic in vivo atlas of human subcortical brain nuclei. Sci Data. 2018:5:180063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Watson C, Petrides M, Rosa M, Tokuno H. The marmoset brain in stereotaxic coordinates. San Diego: Elsevier/Academic Press; 2012. [Google Scholar]
- Pergola G, Danet L, Pitel AL, Carlesimo GA, Segobin S, Pariente J, Suchan B, Mitchell AS, Barbeau EJ. The regulatory role of the human mediodorsal thalamus. Trends Cogn Sci. 2018:22:1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierpaoli C, Walker L, Irfanoglu MO, Barnett A, Basser PJ, Chang L-C, Koay CG, Pajevic S, Rohde G, Sarlls JE, et al. TORTOISE: an integrated software package for processing of diffusion MRI data. International Society for Magnetic Resonance in Medicine (ISMRM). 2010. [Google Scholar]
- Pitkanen A, Amaral DG. Organization of the intrinsic connections of the monkey amygdaloid complex: projections originating in the lateral nucleus. J Comp Neurol. 1998:398(3):431–458. . [DOI] [PubMed] [Google Scholar]
- Price JL, Russchen FT, Amaral DG. The limbic region. II. The amygdaloid complex. In: Bjorkland A, Hokfelt T, Swanson L, editors. Handbook of chemical neuroanatomy. Amsterdam: Elsevier; 1987 [Google Scholar]
- Rempel-Clower NL, Barbas H. Topographic organization of connections between the hypothalamus and prefrontal cortex in the rhesus monkey. J Comp Neurol. 1998:398:393–419. [DOI] [PubMed] [Google Scholar]
- Rijkers K, Temel Y, Visser-Vandewalle V, Vanormelingen L, Vandersteen M, Adriaensens P, Gelan J, Beuls EA. The microanatomical environment of the subthalamic nucleus. Technical note. J Neurosurg. 2007:107(1):198–201. 10.3171/JNS-07/07/0198. [DOI] [PubMed] [Google Scholar]
- Roman E, Weininger J, Lim B, Roman M, Barry D, Tierney P, O'Hanlon E, Levins K, O'Keane V, Roddy D. Untangling the dorsal diencephalic conduction system: a review of structure and function of the stria medullaris, habenula and fasciculus retroflexus. Brain Struct Funct. 2020:225(5):1437–1458. 10.1007/s00429-020-02069-8. [DOI] [PubMed] [Google Scholar]
- Rosene DL, Van-Hoesen GW. The hippocampal formation of the primate brain; a review of some comparative aspects of cytoarchitecture and connections. In: Jones EG, Peters A, editors. Cerebral cortex: further aspects of cortical function, including hippocampus. Vol. 6. New York: Plenum Press; 1987. pp. 345–456, 10.1007/978-1-4615-6616-8_9. [DOI] [Google Scholar]
- Saad ZS, Reynolds RC. Suma. NeuroImage. 2012:62(2):768–773. 10.1016/j.neuroimage.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem KS, Logothetis NK. A combined MRI and histology atlas of the rhesus monkey brain in stereotaxic coordinates. San Diego: Elsevier/Academic press; 2012. [Google Scholar]
- Saleem KS, Price JL, Hashikawa T. Cytoarchitectonic and chemoarchitectonic subdivisions of the perirhinal and parahippocampal cortices in macaque monkeys. J Comp Neurol. 2007:500:973–1006. [DOI] [PubMed] [Google Scholar]
- Saleem KS, Avram AV, Glen D, Yen CC, Ye FQ, Komlosh M, Basser PJ. High-resolution mapping and digital atlas of subcortical regions in the macaque monkey based on matched MAP-MRI and histology. NeuroImage. 2021:245:118759. 10.1016/j.neuroimage.2021.118759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem KS, Avram AV, Yen CC, Magdoom KN, Schram V, Basser PJ. Multimodal anatomical mapping of subcortical regions in marmoset monkeys using high-resolution MRI and matched histology with multiple stains. NeuroImage. 2023:281:120311. 10.1016/j.neuroimage.2023.120311. [DOI] [PubMed] [Google Scholar]
- Sawiak SJ, Shiba Y, Oikonomidis L, Windle CP, Santangelo AM, Grydeland H, Cockcroft G, Bullmore ET, Roberts AC. Trajectories and milestones of cortical and subcortical development of the marmoset brain from infancy to adulthood. Cereb Cortex. 2018:28:4440–4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki F, Hikishima K, Komaki Y, Hata J, Uematsu A, Okahara N, Yamamoto M, Shinohara H, Sasaki E, Okano H. Developmental trajectories of macroanatomical structures in common marmoset brain. Neuroscience. 2017:364:143–156. [DOI] [PubMed] [Google Scholar]
- Setsompop K, Fan Q, Stockmann J, Bilgic B, Huang S, Cauley SF, Nummenmaa A, Wang F, Rathi Y, Witzel T, et al. High-resolution in vivo diffusion imaging of the human brain with generalized slice dithered enhanced resolution: simultaneous multislice (gSlider-SMS). Magn Reson Med. 2018:79:141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklerov M, Dayan E, Browner N. Functional neuroimaging of the central autonomic network: recent developments and clinical implications. Clin Auton Res. 2019:29:555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan H, Baron G, Schwerdtfeger WK. The brain of the common marmoset (Callithrix jacchus). A stereotaxic atlas. Berlin: Springer-Verlag; 1980. [Google Scholar]
- Sternberger LA, Sternberger NH. Monoclonal antibodies distinguish phosphorylated and nonphosphorylated forms of neurofilaments in situ. Proc Natl Acad Sci USA. 1983:80:6126–6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif SD, Mansfield KG, Ratnam R, Ross CN, Ziegler TE. The marmoset as a model of aging and age-related diseases. ILAR J. 2011:52:54–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangavel R, Sahu SK, Van Hoesen GW, Zaheer A. Loss of nonphosphorylated neurofilament immunoreactivity in temporal cortical areas in Alzheimer's disease. Neuroscience. 2009:160:427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier JD, Calamante F, Connelly A. MRtrix: diffusion tractography in crossing fiber regions. Int J Imaging Syst Technol. 2012:22:53–66. [Google Scholar]
- Truong T-K, Darnell D, Song AW. Integrated RF/shim coil array for parallel reception and localized B0 shimming in the human brain. NeuroImage. 2014:103:235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchi J, Chang C, Ye FQ, Russ BE, Yu DK, Cortes CR, Monosov IE, Duyn JH, Leopold DA. The basal forebrain regulates global resting-state fMRI fluctuations. Neuron. 2018:97(4) e944:940–952.e4. 10.1016/j.neuron.2018.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitek JL, Johnson LA. Understanding Parkinson's disease and deep brain stimulation: role of monkey models. Proc Natl Acad Sci USA. 2019:116:26259–26265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells AM, Garcia-Cabezas MA, Barbas H. Topological atlas of the hypothalamus in adult rhesus monkey. Brain Struct Funct. 2020:225:1777–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin CF, Maier SE, Mamata H, Nabavi A, Jolesz FA, Kikinis R. Processing and visualization for diffusion tensor MRI. Med Image Anal. 2002:6:93–108. [DOI] [PubMed] [Google Scholar]
- Woodward A, Hashikawa T, Maeda M, Kaneko T, Hikishima K, Iriki A, Okano H, Yamaguchi Y. The brain/MINDS 3D digital marmoset brain atlas. Sci Data. 2018:5:180009, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa S, Nakamura K, Kohsaka S. Stereotaxic atlas of the marmoset brain with immunohistochemical architecture and MR images. Tokyo, Japan: National Institute of Neuroscience, National Center for Neurology and Psychiatry; 2010 [Google Scholar]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. NeuroImage. 2006:31:1116–1128. [DOI] [PubMed] [Google Scholar]