Abstract
Background
Gastric cancer (GC) is a leading malignant disease in numerous countries, including Taiwan with limited therapeutic options. Animal viruses including oncolytic avian reovirus (ARV) have the possibility to avoid pre-existing immunity in humans, while being safe and immunostimulatory. Here, we provide a novel insight into oncolytic ARV and UV-ARV-sensitized patient’s peripheral blood mononuclear cells (P-PBMCs) and tumor infiltrating lymphocytes (TILs) killing primary GC (PGC) cells through the surface TLR3 and TRAIL/DR4/DR5 immunogenic apoptosis pathway.
Methods
We conducted a comprehensive study to reveal whether ARV- or UV-inactivated ARV (UV-ARV)-modulated P-PBMCs or TILs killing ARV- and UV-ARV-sensitized AGS cells and PGC cells derived from clinical patients and to investigate the regulation of surface TLR3 receptor and upstream signaling pathways. Apoptosis analysis by flow cytometry and Western blot, suppression of signal pathway by specific inhibitors, in situ proximity ligation assay (PLA), time-resolved flurometry and lactate dehydrogenase (LDH) cytotoxicity assays, and an in vitro co-culture model were established to study the interplay between ARV- and UV-ARV-sensitized P-PBMCs and TILs to kill PGC cells and their upstream pathways.
Results
Our results reveal that increased levels of DR4 and DR5 were observed in ARV and UV-ARV sensitized PGC cells through the TLR3/p38/p53 signaling pathway. Importantly, we found that the σC protein of ARV or UV-ARV interacted with surface TLR3 of CD8+ TILs, thereby triggering the TLR3/NF-κB/IFN-γ/TRAIL signaling pathway which induces immunogenic apoptosis of PGC cells. This study sheds further light on the molecular basis behind ARV oncolysis and facilitates the ARV or UV-ARV as a cancer therapeutic.
Conclusions
The study provides novel insights into ARV- or UV-ARV-sensitized P-PBMCs and CD8+ TILs to kill PGC cells through the immunogenic apoptosis pathway. We conclude that P-PBMCs can easily be obtained from GC patients and provide a rich source as TILs to kill PGC cells.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12964-024-01888-0.
Keywords: Apoptosis, Immunogenic apoptosis, Gastric cancer, Immune response, Signal transduction
Background
Oncolytic viruses (OVs) are described as genetically engineered or naturally occurring viruses that specifically replicate and kill cancer cells without affecting normal and healthy cells [1]. OVs including avian reovirus (ARV) are oncolytic viruses that have been extensively studied and applied an oncolytic agent [2–4]. ARVs are not associated with human disease, and pre-existing immunity does not preclude its clinical application [5, 6]. The S1 genome segment of ARV contains three open reading frames that are translated into p10, p17, and σC proteins [7], respectively. The oncolytic potential of ARV was originally thought to be attributed mainly to apoptosis [8–10] and ARV σC is known to be an apoptosis inducer capable of inducing apoptosis in Vero and DF-1 cells through p53 and mitochondria-mediated pathways [9]. The p10 protein of ARV can induce syncytia to facilitate virus spread and distribution within a tumor [8], whereas p17 protein induces autophagy, cell cycle arrest, and host cellular translation shutoff and mediates viral protein synthesis and virus replication [3, 5, 7]. A recent report revealed that p17 protein of ARV induces cell cycle retardation in several cancer cell lines and reduces tumor size in vivo [3].
Cytokine-mediated interactions between immune cells and cancer cells are known to affect various aspects of the tumor microenvironment (TME) [11]. OVs initiate targeted infection and lysis of tumors while expressing therapeutic transgenes such as cytokines, tumor antigens, checkpoint inhibitors in tumors [12]. TLRs and innate immune response pathways initiate pro-inflammatory cascades, culminating in stimulating cytokine production that alter the balance of suppressive and activating immune cells [12, 13]. In response to inflammatory cytokines, TRAIL is secreted, which binds to the surface DR and triggers caspase 3 activation [14]. TRAIL is one of several TNF family members capable of inducing apoptosis through interaction with DR4 and DR5 [14]. ARV-induced cell death may be related to the phenotype of target cells and surrounding TME. PBMCs are blood cells that are an important part of the immune system. They contain a variety of different innate and adaptive cell types [15, 16]. PBMCs in the TME belong to the innate (macrophage/monocyte and NK cells) and adaptive (T and B cells) immune system, and their infiltration of tumors (also called TILs) is highly dependent on the presence of soluble factors in the TME [15, 16]. Although in vitro and in vivo studies suggest that OVs may possess high levels of oncolytic activity [15], potential immunogenicity of ARV is poorly understood. None of studies have directly investigated TRAIL expression on PBMCs and TILs that include the innate and adaptive cellular immune response to oncolytic ARV or UV-ARV. The current study provides a novel insight into oncolytic ARV and UV-ARV-sensitized P-PBMCs and TILs killing PGC cells through the TRAIL/DR4/DR5 immunogenic apoptosis pathway.
Methods
Virus and cell line
The S1133 strain of ARV was used in this study. Human adenocarcinoma gastric cell line (AGS) was cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Biochrom co, Berlin, Germany) supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, and 10 mM 4-(2-hydroxyethyl) piperazine-1-ethanesulphonic acid (HEPES) (pH 7. 2) at 37 °C in a 5% CO2 incubator.
Ethical standards and human samples
Ex vivo normal and malignant gastric tissues were obtained from patients undergoing routine planned cancer-related surgery. Written informed consent was obtained from each patient in accordance with local institutional ethics review and has been approved by the Ethical and Scientific Committee of Taichung Veterans General Hospital (TCVGH-IRB no. SF22141B#1). Clinical characteristics of patient’s samples used in this study are shown in Table 1. All patients have a history of gastric cancer without chemotherapy. The absence of H. pylori infection was confirmed using histological examination. Histological examination of gastric biopsies was obtained from upper gastrointestinal endoscopy which was carried out in all ascertainment of gastric cancer cases.
Table 1.
Clinical characteristics of each sample used in this study
| Pt | Age | Gender | Histopathological diagnosis | Site of origin | Lauren’s classification | Helicobacter pylori |
|---|---|---|---|---|---|---|
| 1 | 60s | F | Poorly differentiated adenocarcinoma | Antrum | Diffuse | Positive |
| 2 | 50s | M | Moderately to poorly differentiated adenocarcinoma | Greater curvture | Mixed | Negative |
| 3 | 70s | F | Moderately differentiated adenocarcinoma | Angular incisure | Instestinal | Negative |
| 4 | 40s | M | Moderately to poorly differentiated adenocarcinoma | Antrum | Instestinal | Negative |
| 5 | 60s | F | Moderately differentiated adenocarcinoma | Angular incisure | Instestinal | Positive |
| 6 | 60s | M | Poorly differentiated adenocarcinoma | Greater curvture | Instestinal | Negative |
| 7 | 50s | F | Poorly differentiated adenocarcinoma | Body | Diffuse | Negative |
| 8 | 60s | M | Poorly differentiated adenocarcinoma | Lesser curvture | Diffuse | Negative |
| 9 | 40s | F | Moderately differentiated adenocarcinoma | Lesser curvture | Instestinal | Negative |
| 10 | 50s | M | Poorly differentiated adenocarcinoma | Greater curvture | Diffuse | Negative |
| 11 | 50s | M | Enteroblastic differentiation | Lesser curvture | Not defined | N/A |
| 12 | 50s | F | Poorly differentiated adenocarcinoma | Antrum | Diffuse | Negative |
| 13 | 60s | M | Moderately differentiated adenocarcinoma | Antrum | Instestinal | Negative |
| 14 | 60s | M | Poorly differentiated | Cardiac | Diffuse | Negative |
| 15 | 70s | F | Moderately differentiated adenocarcinoma | Cardia | Instestinal | Negative |
| 16 | 60s | F | Moderately differentiated adenocarcinoma | Lesser curvture | Intestinal | Negative |
| 17 | 70s | M | Moderately to poorly differentiated adenocarcinoma | Antrum | Intestinal | Positive |
| 18 | 60s | M | Poorly differentialted adenocarcinoma | Angular incisure | Diffuse(or mixed) | Negative |
| 19 | 50s | M | Moderately differentiated adenocarcinoma | Antrum | Diffuse | Positive |
| 20 | 70s | M | Moderately to poorly differentiated adenocarcinoma | Cardia | Diffuse | Negative |
| 21 | 40s | F | Moderately differentiated adenocarcinoma | Cardia | Intestinal | Negative |
Pt: Patient; Helicobacter pylori: Histological identification
N/A: not available
Electrophoresis and Western blot assays
Cells were seeded in 6-well cell culture dishes one day before infecting with virus or transfecting with plasmid as described above. Collected cells were washed twice with 1X PBS and lysed with lysis buffer (Cell Signaling, St. Louis, USA). The cell lysates were determined the concentration of solubilized protein with the Bio-Rad protein assay (Bio-Rad Laboratories, St. Louis, USA). Assays were performed according to the manufacturer’s protocol. Equal amounts of samples were mixed with 2.5X Laemmli loading buffer and boiled for 10 min in a water bath. The samples were analyzed in a 12% sodium dodecyl sulphate (SDS)-polyacrylamide gel electrophoresis (PAGE) gel and transferred to PVDF membrane (GE Healthcare Life Sciences, Chicago, USA). Protein expression was examined using the respective primary antibody and horseradish peroxidase (HRP)-conjugated secondary antibody conjugate. The results were detected on X-ray films (Kodak, Rochester, USA) after the membrane incubation with enhanced chemiluminescence (ECL plus) (Amersham Biosciences, Little Chalfont, England).
Apoptosis assays
Apoptosis was assessed by flow cytometric analysis of cells stained with annexin V-fluorescein isothiocyanate (FITC) and PI according to manufacturer’s instruction (PI/annexin V-FITC Cell Apoptosis Detection kit, Elabscience, Houston, USA). PBMCs and AGS co-culture cells were seeded at 106 cells/mL in 6-well plates for 24 h followed by inoculation of ARV at an MOI of 10, UV-ARV (MOI = 10 or MOI = 100), or medium alone in 24 h and 48 h, respectively. Briefly, cells were washed and resuspended in 500 µL PBS followed incubation with 10 µL PI and 10 µL annexin V-FITC at room temperature in the dark for 15 min and analyzed by flow cytometry.
Human primary gastric cell culture from fresh surgical gastric tissues
The specimens are collected in Dulbecco’s modified Eagle’s medium (DMEM) (Biochrom Co, Berlin, Germany) containing 1% Penicillin/Streptomycin for transport to our laboratory. Cell culture of primary human gastric cancer cells and gastric normal epithelial cells were purified and maintained in DMEM medium supplemented with 10% FBS as previously described [17, 18]. On the next day, cell culture was rinsed with PBS twice to remove non-adherent cells. The medium was changed every 3–7 days, depending on the density of cell growth. The colonies increase in size and spread out, resulting in some cells separating at the periphery of the colonies after 2 weeks of culture. Gastric normal epithelial cells were confirmed by flow cytometry analysis of cytokeratin 18 (CK-18) expression. Primary GC cells were identified using granulin (GRN) markers [17]. These primary cells were maintained in culture for up to 4–8 weeks.
Sorting of T cells, B cells, monocytes/macrophages, NK Cells and PGC cells
PBMCs were stained with the CD3 Ab for T cells, CD56 Ab for NK cells, CD19 Ab for B cells, CD14 Ab for monocyte/macrophage cells. TILs were stained with CD8 Ab for cytotoxic T cells (CTLs), CD4 Ab for helper T cells (Th cells), CD56 Ab for NK cells, and CD14 Ab for monocyte/macrophage cells. Gastric normal epithelial cells were stained with CK-18. PGC cells were stained with GRN. Sample acquisition and cell sorting was managed on the BD FACSMelody™ cell sorter (BD Biosciences, San Jose, USA) and BD Chorus software (BD Biosciences, San Jose, CA). All antibodies used in this study are shown in Table S1.
Isolation of TILs
The generation of TIL cultures by tumor has been described in detail [19]. Briefly, the tumor removed from cancer patients was placed on a plate with 5% FBS in Hank’s Balanced Salt Solution (HBSS) buffer (Gibco, New York, USA) on ice and disintegrated using scissors. The homogenate was collected and treated with 1 mg/ml type IV collagenase (Sigma, St. Louis, USA) and 0.05 mg/ml DNase (Promega, Madison, USA) for 30 min at 37 °C with gentle agitation. The digested extract was screened using a 100-mesh, and the filtrate was washed with 5% FBS in HBSS buffer and centrifuged at 600xg for 7 min at 4 °C. The cell pellet obtained was treated with ACK erythrocyte lysis buffer (155 mM NH4Cl, 10 mM KHCO3, and 1 mM Na2EDTA, pH 7.3) for 5 min at room temperature. Finally, TILs were resuspended in RPMI 1640 medium with 10% FBS. Cells were harvested after 7 to 14 days of culture. Each initial well was considered to be an independent TIL culture and maintained separately from the others.
Co-culturing of immune cells and cancer cells in presence of ARV or UV-ARV
The ratio of PBMCs (effector cells) to AGS cells (cancer cell lines) is 5:1. The selected ratio is according to a previous report by Doumba [20]. For direct in vitro co-culture, PGC cells were plated with p-PBMCs or TILs from the same patient at a 1:5 ratio in reduced-serum medium (2% FBS). The selected ratio is based on AGS cells and PBMCs. The cultures were incubated for 3 days, after which cells were sensitized with the ARV or UV-ARV for 24 h and 48 h.
Detection of cytotoxicity of PBMCs and TILs
The cytotoxicity of PBMCs and TILs were estimated by quantification of LDH activity in the culture medium by using the QuantiChrom™ LDH Cytotoxicity Assay Kit (BioAssay Systems, Hayward, USA) [21, 22]. Briefly, cytotoxicity assays were carried out in 96-well plates with a final sample volume of 100 µl/well. Target cells (AGS cells and PGC cells, 2 × 105/ml cells) in 50 µl/well were co-cultured with effector cells (normal PBMCs, patient’s PBMCs, and TILs) at various effector to target ratios (5:1) for 4 h [23].
ARV or UV-ARV sensitized-CD8+TILs mediated cytotoxicity assay using time-resolved fluorometry
ARV or UV-ARV sensitized-CD8+TILs cytotoxicity was determined using the DELFIA® EuTDA Cytotoxicity Reagents (PerkinElmer Life Sciences, Waltham, MA, USA). Target cells (PGC cells) were incubated with freshly prepared 10 µM BATDA (a fluorescence enhancing ligand) in 2 ml of culture medium for 30 min at 37 °C, and washed. Next, 100 µl of BATDA-labeled target cells (PGC cells) were transferred into a round bottom sterile plate and co-cultured with ARV or UV-ARV sensitized-CD8+TILs at effector/target ratios was 5:1. After incubation, 20 µl of supernatant from each well was transferred to the wells of flat-bottom 96 well plates. 180 µl of europium (Eu) solution was then added to form highly fluorescent and stable chelates (EuTDA), and the fluorescence of these chelates were measured by time resolved fluorometry (Enspire 2300-0000, PerkinElmer). The percent of specific release was calculated using (experimental release – spontaneous release)/ (maximum release – spontaneous release) X 100(%). All experiments were performed in triplicate.
Analysis of P-PBMCs or TILs including CD3+, CD8+, CD56+, CD19+, CD14+, and TRAIL by flow cytometry
After washing three times with PBS buffer, 105-106 cells of P-PBMCs or TILs were divided into 1.5 cc centrifuge tubes. The CD3, CD56, CD19, and CD14 conjugated fluorescent antibodies were added and incubated at dark at 4°C for 30 min. After washing once, the cells were resuspended in PBS. TRAIL expression levels on P-PBMCs or TILs after ARV or UV-ARV sensitization were analyzed. TRAIL on CD8+ or CD3+, CD14+, CD19+, and CD56+ cells was analyzed 24 h post treatments using flow cytometry. Data acquisition and analysis were performed using a BD FACS Canto II flow cytometer with CellQuest™ software (Becton-Dickinson, Mississauga, USA). Representative results are shown in histograms based on 104 gated cells in all conditions, and cell viability was > 95%, as assessed by propidium iodide (PI) exclusion.
Suppression of TLR3 and upstream signaling by inhibitors
The following compounds were used to restore intracellular signaling: the p53 inhibitor Pifithrin-α p53 inhibitor (PFT-α) (20 µM), the p38 inhibitor (SB202190) (20 µM), the CU-CPT 4a (TLR3-IN-1) TLR3 signaling inhibitor (10 µg/mL). Inhibitory compounds-induced cell toxicity was assessed through analyzing the level of cell death by flow cytometry. The level of concentration that was not cytotoxic and carry inhibitory effect were used in all experiments.
Proximity ligation assay (PLA)
In this work, interaction between ARV σC and surface TLR3 on CD8+ TILs or PGC cells were analyzed by in situ PLA using the commercial kit Duolink (Sigma-Aldrich, cat no. DUO 92008) according to manufacturer’s instructions. PLA allows the detection of direct protein-protein interactions at distances of < 40 nm in intact fixed cells. Briefly, CD8+TILs or primary GC cells preincubated with ARV at a MOI of 10 or UV-ARV (100 MOI), and, after 30 min of incubation, fixed with 4% paraformaldehyde for 10 min at room temperature. After incubation with a blocking buffer for 30 min at 37 °C, cells were incubated with the respective antibodies as primary antibodies. After several washes, cells were incubated with the PLA probes linked, anti-mouse, and anti-rabbit secondary antibodies. Finally, the cells were subjected to ligation and amplification reactions. The PLA signal was detected in a confocal microscope (Inverted Olympus MPhot).
Phosphoprotein staining [24]
Phosphoproteins were measured in either unstimulated PGC cells or stimulated with ARV or UV-ARV for 24 h in the presence or absence of inhibitors. For anti-p38, anti-phospho-p38, anti-p53 and anti-phospho-p53 antibody detection, the intracellular staining was performed using Fixation/Permeabilization Solution Kit (Cytofix/Cytoperm BD Biosciences, San Jose, USA), according to the manufacturer’s instructions. Data were collected with a FACSCANTO II multicolor flow cytometer and analyzed.
Statistical analysis
Statistical analyses and figures were generated using GraphPad Prism 8.0 software (GraphPad Software Inc., La Jolla, USA). Differences between means were evaluated using the Student’s t-test and were deemed significant at *p ≤ 0.05 and **p ≤ 0.01.
Results
ARV-induced apoptosis by TRAIL in AGS cells and PGC cells derived from clinical patients
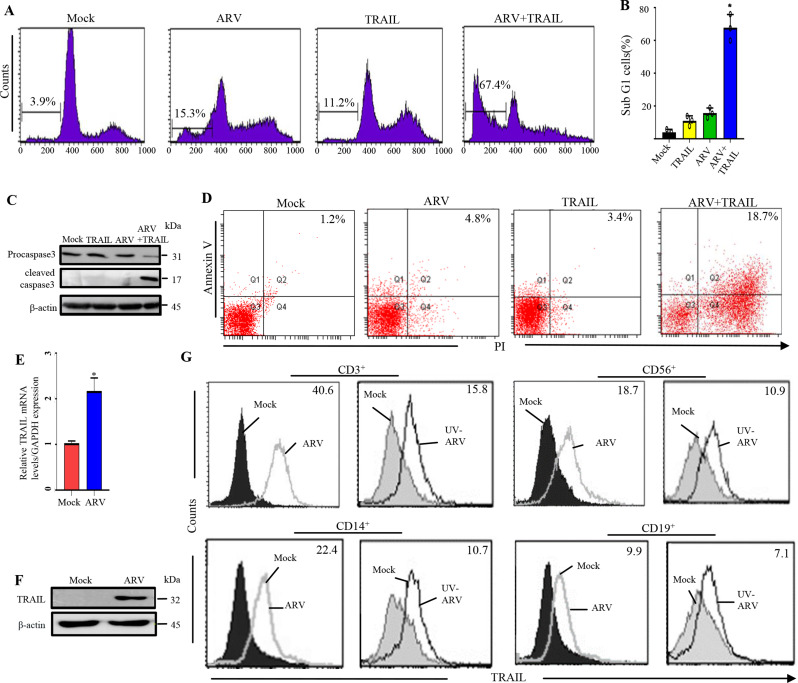
In this work, virus titers and the levels of DR4, DR5, TLR3, and cytokines were analyzed in ARV-infected AGS cells (Fig. S1-2). The increase in T helper(Th)-1 cytokines (IFN–γ and IL-12) observed in ARV-infected AGS cells (Fig. S2C-D), which can indicate an activation tendency towards Th1 cells. To investigate whether TRAIL is involved in ARV-induced AGS cell apoptosis, ARV-induced AGS cell apoptosis by TRAIL was examined. For this purpose, AGS cells were infected with ARV at an MOI of 10 for 24 h, followed by analysis of sub-G1 population by flow cytometry. Importantly, in the presence of TRAIL, ARV significantly enhanced the percentage of sub-G1 population (from 3.9 ± 0.6% to 67.4 ± 7.2%), accompanied by the increased levels of cleaved caspase 3 which was detected by Western blotting (Fig. 1C) while TRAIL alone only increased to 11.2 ± 1.6% percentage (Fig. 1A and B). Annexin V and PI double staining were also used to examine ARV-induced AGS cell apoptosis. The flow cytometry data was plotted in two-dimensional dot plots where PI represented versus annexin V-FITC. Apoptotic cells which are PI and annexin double positive (PI/FITC +/+) were shown in Fig. 1D. These findings demonstrate that ARV induces AGS cell apoptosis through the TRAIL/DR4/DR5 apoptotic pathway. Thus, we next intended to examine TRAIL expression in human PBMCs after ARV sensitization. TRAIL levels were analyzed in PBMCs incubated with ARV for 24 h and found that ARV upregulated expression levels of TRAIL in PBMCs (Fig. 1E and F). Therefore, PBMCs were examined by two-color flow cytometry 24 h post ARV sensitization. A significant increase in TRAIL levels was observed post-sensitization in all four major PBMCs populations. Importantly, we found that higher expression levels of TRAIL were observed in CD3+ cells (Fig. 1G). We next confirmed whether UV-ARV can trigger TRAIL expression in PBMCs. For this purpose, the TRAIL expressions of CD56+, CD14+, CD3+ and CD19+ were examined by two-color flow cytometry 24 h after UV-ARV incubation. Interestingly, it was observed that stimulation with UV-ARV was sufficient to induce TRAIL expression on PBMCs (Fig. 1G). Taken together, our findings revealed that TRAIL expression is not dependent on direct infection of the TRAIL-expressing cells (Fig. 1G).
Fig. 1.
ARV-induced apoptosis in AGS cells through the TRAIL signaling pathway and ARV-induced expression of TRAIL on PBMCs driven by IFN-γ sensitization. (A) AGS cells were infected with ARV at an MOI of 10 for 24 h and sensitized in the presence or absence of recombinant TRAIL protein (25 ng/mL). Sub-G1 cell populations were analyzed by flow cytometry. Counts: the number of events (cell count) on the y-axis. (B) Graph shown represents the mean ± SE calculated from three independent experiments. *p < 0. 05 * * p < 0.01. In this work, the statistical methods of Figs. 2, 3, 4, 5, 6, 7 and 8 are the same as the Fig. 1. (C) AGS cells were infected with ARV at an MOI of 10 for 24 h and sensitized in the presence or absence of recombinant TRAIL protein (25 ng/mL). Cell lysates were analyzed by Western blot assays. All original/uncropped blots and images from this study are provided in supplementary Fig. 7. (D) To detect cell death, annexin V and PI double staining was used in flow cytofluorimetric analyses. The data generated by flow cytometry are plotted in two-dimensional dot plots in which PI is represented versus annexin V-FITC. Apoptotic cells which are PI and Annexin positive (PI/FITC +/+). PBMCs were sensitized with ARV at an MOI of 10. TRAIL levels were analyzed 24 h post-sensitization. PBMCs were isolated from normal healthy volunteers (n = 3). Similar results were observed in 3 different PBMC samples. The expression levels of TRAIL were examined by qRT-PCR (E) Western blot assays (F). (G) TRAIL expression on human PBMC after ARV or UV-ARV sensitization. The expression levels of TRAIL were analyzed by 24 h later on CD3+, CD14+, CD19+, and CD56+ cells using two-color flow cytometry. Representative results are shown in histograms based on 104 gated cells in all conditions, relative mean fluorescence intensity (RMFI) is shown on histograms. Cell viability was > 95%, as assessed by PI exclusion. Similar results were observed using at least 3 different PBMC donors
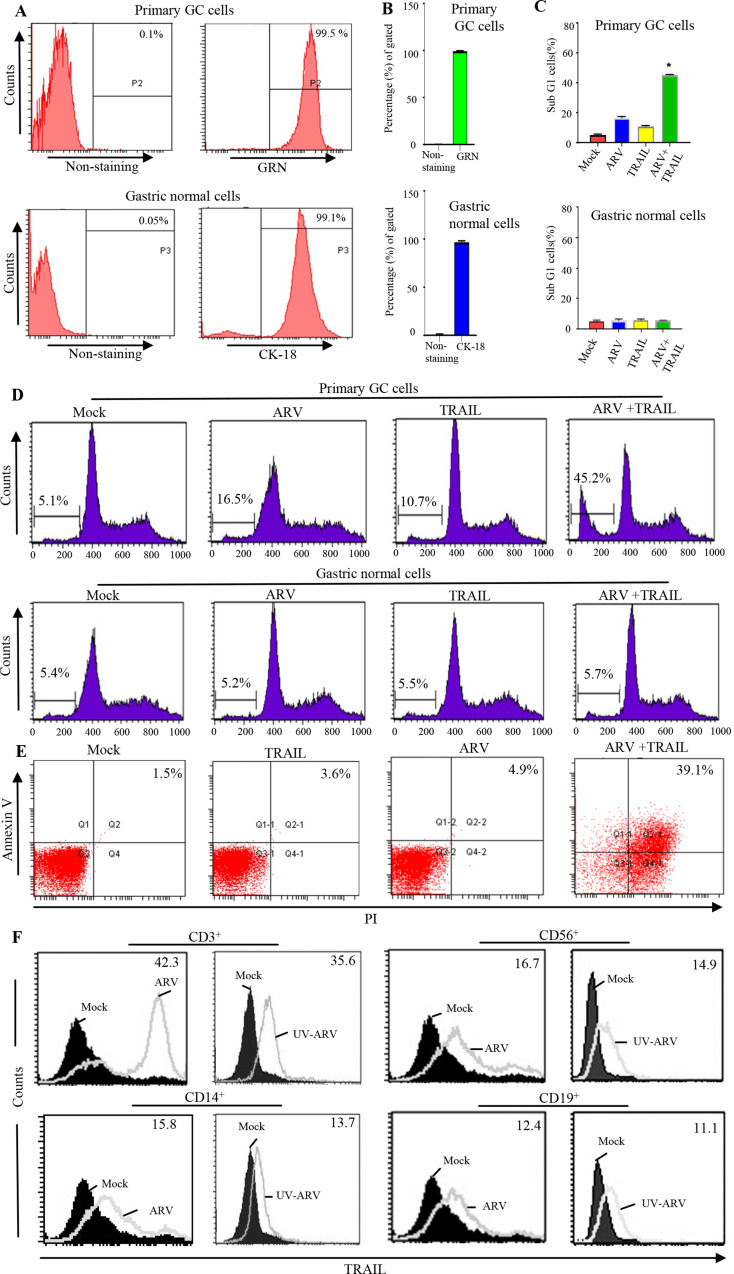
Our findings that ARV-induced apoptosis by TRAIL in AGS cells but not PBMCs are shown in the study details of supplementary information (Fig. S3-S5). Thus, we next wanted to study whether the same effect is also achieved on the PGC cells and P-PBMCs of gastric cancer patients. In this study, normal patient epithelial cells and PGC cells were characterized by the detection of cytokeratin 18 antigen and GRN markers, respectively (Fig. 2A and B). Next, sub-G1 populations were analyzed by flow cytometry where PGC cells were infected with ARV with an MOI of 10 for 24 h. In the presence of TRAIL, ARV significantly enhanced the percentage of sub-G1 populations, while TRAIL alone only slightly induced apoptosis (Fig. 2C and D). These results suggest that ARVs induce apoptosis in PGC cells through the TRAIL apoptotic signaling. ARV-sensitized P-PBMCs can selectively and efficiently kill malignant PGC cells sparing normal counterparts (Fig. 2C and D). Annexin V and PI double staining were used to examine ARV-induced apoptosis in PGC cells. The data generated by flow cytometry was plotted in two-dimensional dot plots and analysis indicated that apoptotic cells were PI and annexin double positive (PI/FITC+/+) (Fig. 2E). P-PBMCs were isolated from volunteers of GC patients.
Fig. 2.
ARV-induced apoptosis in PGC cells through the TRAIL signaling. (A and B) To confirm the normal epithelial cells, cytokeratin 18 (CK-18) staining was performed. The highly positive staining for granulin (GRN) discriminated the PGC cells from normal gastric cells. (C and D) PGC cells were infected with ARV at an MOI of 10 for 24 h and sensitized in the presence or absence of recombinant TRAIL protein (25 ng/mL). Sub-G1 cell populations were analyzed by flow cytometry. (E) To detect cell death, annexin V/PI double staining was used in flow cytofluorimetric analyses. The data generated by flow cytometry are plotted in two-dimensional dot plots in which PI is represented versus annexin V-FITC. Apoptotic cells which are PI and annexin positive (PI/FITC +/+). TRAIL expression on P-PBMCs after ARV and UV-ARV sensitization. (F) The expression levels of TRAIL were analyzed 24 h later on CD3+, CD14+, CD19+, and CD56+ cells using two-color flow cytometry, relative mean fluorescence intensity (RMFI) is shown on histograms. Representative results are shown in histograms based on 104 gated cells in all conditions, and cell viability was > 95%, as assessed by PI exclusion. Similar results were observed using at least 3 different P-PBMCs donors
ARV and UV-ARV- sensitized TRAIL expression on GC patient’s PBMCs
TRAIL levels were analyzed in P-PBMCs sensitized with ARV for 24 h. P-PBMCs were examined by two-color flow cytometry 24 h post ARV sensitization. When sensitized, a significant TRAIL levels were observed in all four major P-PBMC populations (Fig. 2F). We found that highest expression levels of TRAIL were observed in patient’s CD3+ T cell (Fig. 2F). We next wanted to confirm whether UV-ARV can trigger TRAIL expression of P-PBMCs. Thus, CD56+, CD14+, CD3+, and CD19+ were examined by two-color flow cytometry for TRAIL expression 24 h after UV-ARV sensitization. Interestingly, sensitization with UV-ARV was sufficient to induce TRAIL expression on P-PBMC populations (Fig. 2F). Taken together, our results revealed that TRAIL expression is independent of direct infection.
TRAIL upregulation dependent on IFN-γ sensitization but not direct infection and IFN-γ driven expression of TRAIL on P-PBMCs
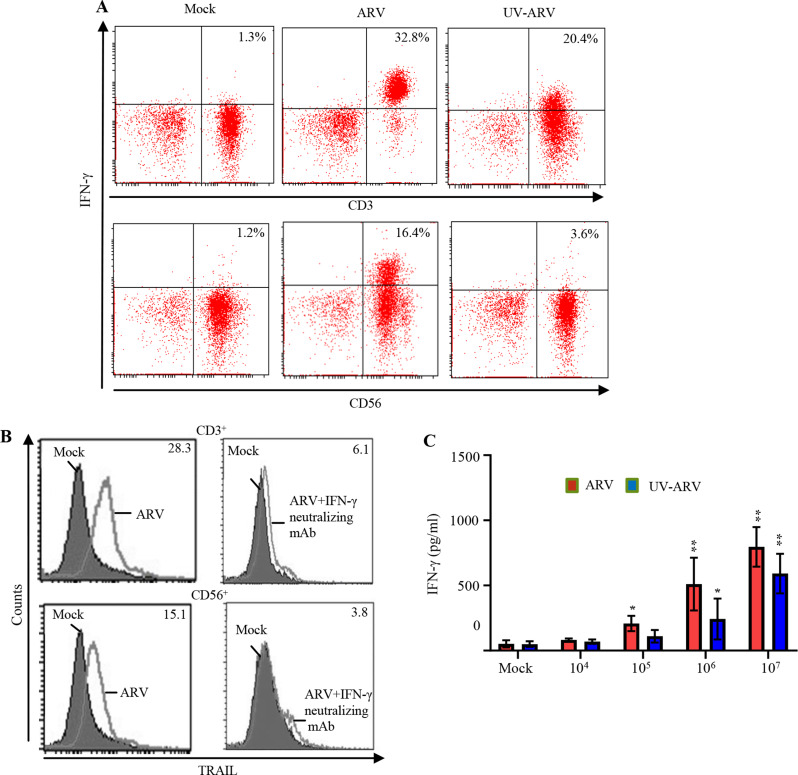
Our analysis of IFN-γ levels in ARV-sensitized P-PBMCs revealed that ARV-sensitized P-PBMCs produce high levels of IFN-γ (Fig. 3A). A similar trend was also observed in UV-ARV-sensitized P-PBMCs (Fig. 3A). Furthermore, intracellular staining of IFN-γ revealed that CD3+ and CD56+ cells produced a high level of IFN-γ cytokine after ARV sensitization (Fig. 3A). To verify that TRAIL expression was driven by IFN-γ, the expression levels of TRAIL in ARV-sensitized cells in the presence of anti-IFN-γ neutralizing antibodies or medium alone were analyzed. The increased levels of TRAIL in CD3+ and CD56+ cells were observed in ARV-sensitized P-PBMCs. This effect was reversed in cells-treated with the IFN-γ antibody (Fig. 3B). Taken together, the results demonstrate that increased levels of TRAIL on ARV-sensitized P-PBMCs are regulated by the IFN-γ signaling. To investigate whether ARV or UV-ARV sensitizes IFN-γ of P-PBMCs, the ARV or UV-ARV-sensitized P-PBMCs were divided into cultures with decreased cell numbers. As expected, decreased numbers of P-PBMCs accompanied by decreased levels of IFN-γ in the cultures (Fig. 3C). These finding demonstrates that increased levels of TRAIL on ARV-sensitized or UV-ARV- sensitized P-PBMCs are regulated by IFN-γ signaling.
Fig. 3.
ARV or UV-ARV upregulates the IFN-γ expression levels in P-PBMCs. (A) IFN-γ expression by P-PBMCs after ARV or UV-ARV stimulation. P-PBMCs were sensitized with ARV or UV-ARV. Intracellular IFN-γ levels were analyzed 24 h post treatment later in CD3+ and CD56+ cells using two-color flow cytometry. (B) P-PBMCs cultured with 5 µg/ml anti-IFN-γ Ab or isotype control Ab for 1 h followed by sensitization with ARV and cultured for 24 h. Representative results for CD3+ and CD56+ cells are shown in histograms based on at least 104 gated cells. RMFI is shown on histograms. Similar results were observed using 3 different P-PBMC donors. (C) Decreasing numbers of P-PBMCs (ranging107to104) were stimulated with ARV or UV-ARV, respectively. After stimulated, IFN-γ levels in the culture supernatants were determined by ELISA
ARV or UV-ARV-sensitized GC patient’s PBMCs killing ARV-infected or UV-ARV sensitized PGC cells
To investigate whether sensitization of P-PBMCs kill PGC cells, we examined responsiveness of PGC cells co-cultured with P-PBMCs after ARV or UV-ARV sensitization. ARV-unsensitized P-PBMCs induces minimal apoptosis of PGC cells, whereas ARV or UV-ARV-sensitized P-PBMCs induced strong apoptosis of PGC cells (Fig. 4A-B). To confirm whether ARV-modulated cytotoxic activity of P-PBMCs is TRAIL-dependent, PGC cells co-cultured with P-PBMCs were treated with either DR5:Fc or Fas: Fc prior to their sensitization with ARV or UV-ARV. Under these conditions, DR5:Fc reversed apoptosis of PGC cells, whereas no change was observed in ARV-sensitized P-PBMCs treated with Fas: Fc (Fig. 4A-B). Similar results were observed in UV-ARV-sensitized P-PBMCs (Fig. 4A-B), indicating that infection is not required to induce TRAIL expression in P-PBMCs. UV-ARV potently activate P-PBMCs to induce apoptosis of PGC cells (Fig. 4A-B). Furthermore, the cytotoxic effect on PGC cells was assessed by an LDH release assay. As shown in Fig. 4C, after sensitization with ARV or UV-ARV, CD3+ cells displayed a strong cell killing activity on PGC cells.
Fig. 4.
ARV- and UV-ARV-sensitized P-PBMCs-dependent cytolysis of PGC cells (PGCs). (A) P-PBMCs were co-cultured with PGCs followed by sensitization with ARV for 24 h. The ratio of cell numbers of P-PBMCs (effector cells) and PGCs (target cells) was 5:1. DR5:Fc (20 µg/ml) was used to inhibit ARV-sensitized P-PBMCs killing PGCs. Fas: Fc (20 µg/ml) was used as a control negative. Cell death was measured by SubG1 (A) and Annexin V/PI (B). Data represent the mean of triplicate experiments, and experiments were repeated at least three times using different donor P-PBMCs with similar results. (C) P-PBMCs were co-cultured with PGCs followed by sensitization with ARV or UV-ARV for 24 h and 48 h, respectively. The ratio of coculture cell numbers of P-PBMCs and PGCs was 5:1. Cell death was measured by LDH cytotoxicity assay. Data represent the mean of triplicate experiments, and experiments were repeated at three times using different donor P-PBMCs with similar results
ARV or UV-ARV-sensitized CD8+ TILs but not CD4+ TILs killing PGC cells
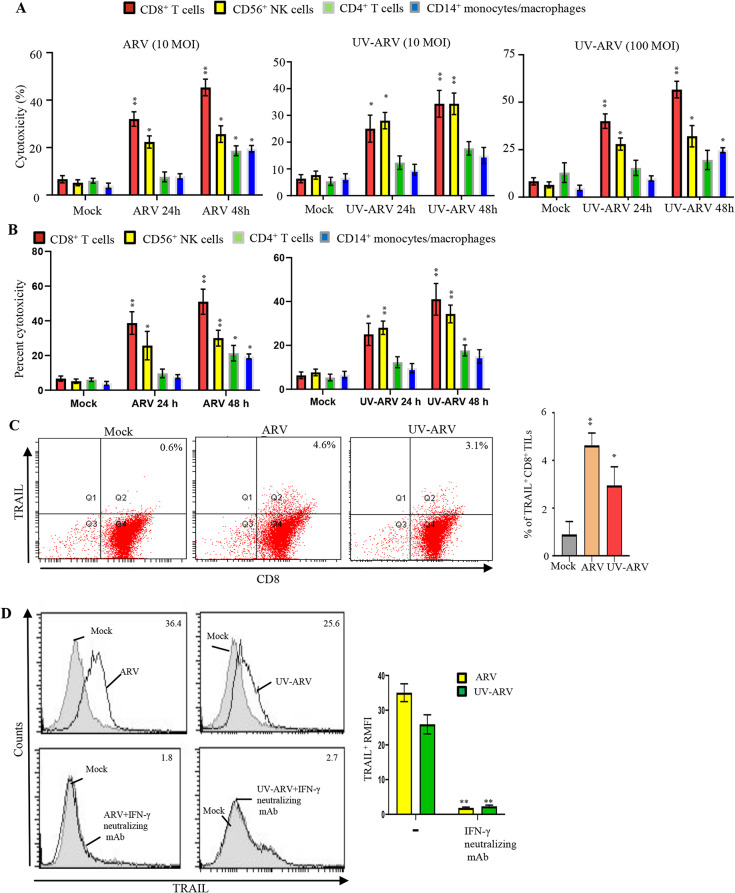
We found that gastric TILs are composed of CD4+ and CD8+ (about 75%), CD14+ (< 10%), and CD56+ (< 5%) infiltrating the gastric tumor together. Compared with CD8+ TILs, CD4+ TILs were more efficient at host immune activation but less capable of direct tumor killing. Since CD8+ TILs maintain high cytotoxicity, cytotoxic activity on PGC cells was assessed by LDH release assay. As shown in Fig. 5A, CD8+ TILs display a strong cell killing activity on PGC cells when sensitized with ARV (10 MOI) or UV-ARV (10 and 100 MOIs). Treatment of CD8+ TILs with ARV (10 MOI ) or UV-ARV at various MOIs (10–100) could not induce apoptosis in CD8+TILs (fig. S6). CD8+ TILs cytotoxicity was also assessed using DELFIA EuTDA cell cytotoxicity assays. Tumor cells (PGC cells) were labeled with BATDA (a fluorescence enhancing ligand). Subsequently, ARV or UV-ARV-sensitized CD8+ TILs were added to the BATDA labeled PGC cells. ARV or UV-ARV-unsensitized CD8+ TILs showed minimal cytotoxicity of PGC cells, whereas ARV or UV-ARV-sensitized CD8+ TILs induced strong cytotoxic effect of PGC cells. As shown in Figure Fig. 5B, ARV or UV-ARV-sensitized CD8+ TILs displayed strong cell killing activity on PGC cells. TRAIL expression by CD8+ TILs sensitized with ARV or UV-ARV were shown in Fig. 5C. To further confirm the necessity of IFN-γ to drive TRAIL expression, CD8+ TILs were sensitized with ARV or UV-ARV for 24 h followed by treatments with neutralizing IFN-γ mAb or isotype mAb. Our results revealed that treatment with the neutralizing IFN-γ mAb failed to induce TRAIL expression (Fig. 5D), suggesting that TRAIL on ARV or UV-ARV-sensitized TILs is induced by IFN-γ. IFN-γ produced by CD8+ TILs enhanced TRAIL expression was essential to sustaining the cytotoxicity of CD8+ TILs. After treatment of ARV or UV-ARV, we found that the expression level of TRAIL was increased in ARV- or UV-ARV-sensitized CD8+ TILs thereby enhancing TRAIL-specific killing PGC cells. This is the first report to show direct interaction between CD8+ TILs and PGC cells regulated by ARV and UV-ARV in an in vitro co-culture system.
Fig. 5.
ARV or UV-ARV induces most immunogenic apoptosis in PGC cells (PGCs) and upregulates the TRAIL expression levels by CD8+TILs. CD8+T cells, CD4+ T cells, CD56+NK cells, and CD14+monocyte/macrophages were co-cultured with PGCs followed by sensitization with ARV or UV-ARV for 24 h and 48 h, respectively. The ratio of coculture cell numbers of TILs and PGCs was 5:1. Cell death was measured by LDH cytotoxicity assay (A) and DELFIA EuTDA cytotoxicity detection (B). Data represent the mean of triplicate experiments, and experiments were repeated at three times using different donor TILs with similar results. (C) The expression levels of TRAIL were analyzed on CD8+ TILs 24 h post treatment ARV or UV-ARV using two-color flow cytometry. Cell viability was > 95%, as assessed by PI exclusion. Similar results were observed using at least 3 different CD8+ TILs donors. (D) CD8+TILs cultured with 5 µg/ml anti-IFN-γ Ab or isotype control Ab for 1 h, followed by sensitization with ARV or UV-ARV and cultured for 24 h. Representative results for CD8+ TILs are shown in histograms, RMFI is shown on histograms. Similar results were observed using 3 different CD8+ TILs donors
ARV- or UV-ARV-sensitized CD8+TILs expressing TRAIL which kills PGC cells
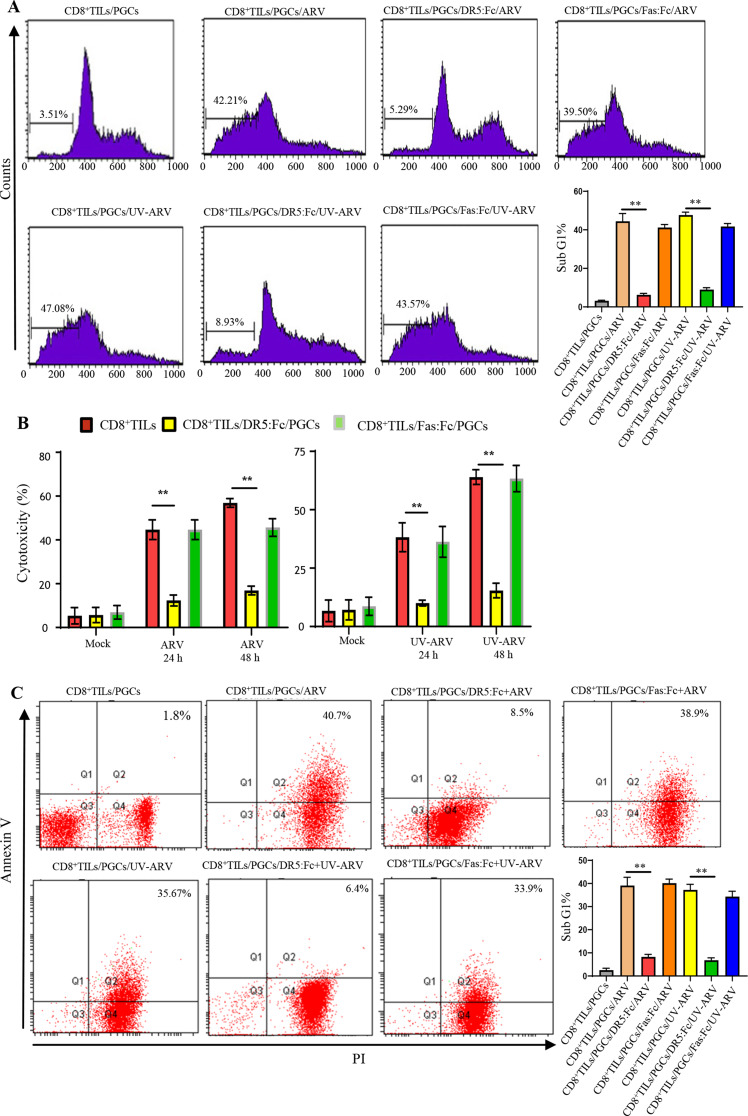
To investigate whether sensitization of CD8+ TILs directly kill PGC cells in TME, we examined responsiveness of PGC cells co-cultured with CD8+ TILs after different treatments with ARV or UV-ARV. ARV- or UV-ARV-unsensitized CD8+ TILs induced minimal apoptosis of PGC cells, whereas ARV or UV-ARV-sensitized CD8+ TILs induced strong apoptosis of PGC cells. To further confirm whether ARV- or UV-ARV-modulated cytotoxic activity of CD8+ TILs is through a TRAIL-dependent manner, PGC cells co-cultured with CD8+ TILs were treated with either DR5:Fc or Fas: Fc prior to their sensitization with ARV or UV-ARV (Fig. 6A-C). Under these conditions, DR5:Fc reversed apoptosis of PGC cells, while no change was observed in UV-ARV or ARV-sensitized CD8+ TILs treated with Fas: Fc (Fig. 6A-C). Similar results were observed in UV-ARV-sensitized CD8+ TILs (Fig. 6A-C), indicating that ARV infection is not required to induce TRAIL expression in CD8+ TILs. Our study documented that ARV- and UV-ARV-sensitized CD8+ TILs killing PGC cells was mainly mediated by IFN-γ and TRAIL. In vitro co-cultures revealed that killing of PGC cells were enhanced by ARV- or UV-ARV-sensitized CD8+ TILs.
Fig. 6.
ARV- or UV-ARV-sensitized CD8+TILs expressing TRAIL which kills PGC cells (PGCs). (A) CD8+TILs were co-cultured with PGCs followed by sensitization with ARV or UV-ARV for 24 h, respectively. The ratio of cell numbers of CD8+TILs (effector cells) and PGCs (target cells) was 5:1. DR5:Fc (20 µg/ml) was used to inhibit ARV or UV-ARV-sensitized CD8+TILs killing PGCs. Fas: Fc (20 µg/ml) was used as a control negative. Cell death was measured by Sub-G1 (A), LDH cytotoxicity assay (B), and Annexin V/PI (C). Similar results were observed using 3 different CD8+TILs donors. (D) PGCs were sensitized with UV-ARV (100 MOI) or ARV (10 MOI) for 24 h. Cell-surface DR4 and DR5 were analyzed by flow cytometry
ARV or UV-ARV-sensitized CD8+ TILs induces the IFN-γ expression through the TLR3/NF-κB signaling pathway
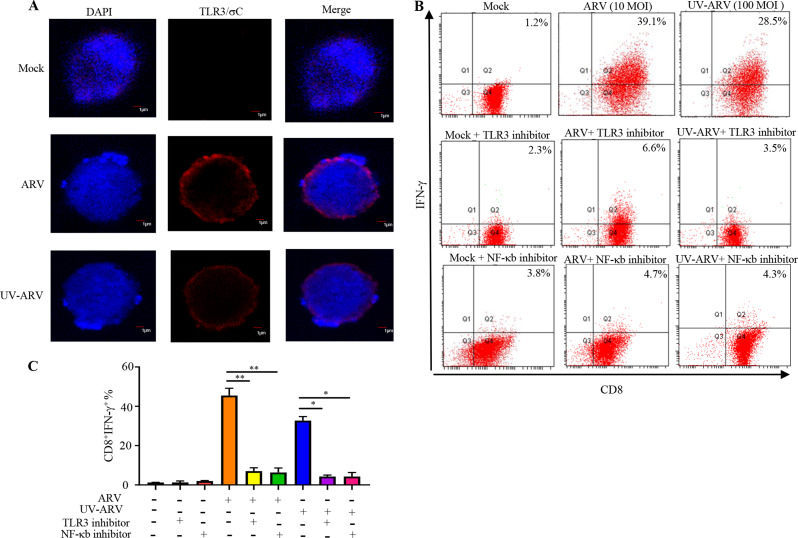
A previous study suggested that human effector CD8+ cells express TLR3 as a functional coreceptor [25]. To determine whether ARV or UV-ARV σC protein interacts with cell surface TLR3 on CD8+ TILs, interactions between σC protein with cell surface TLR3 of CD8+ TILs were analyzed by in situ PLA. Tumor infiltrating cytotoxic T-cells carry higher nuclear to cytoplasmic ratios [26] and oncolytic viruses specifically replicate and infect cancer cells without affecting healthy cells including CD8+ TILs [1, 15]. Our results clearly indicated that σC protein interacts with cell surface TLR3 of CD8+ TILs (Fig. 7A). In contrast, no signal was observed in negative controls (Fig. 7A). Previous study had indicated that TLR3-induced signaling spreads to several adaptors and downstream activation of NF-κB [25]. These prompted us to investigate whether ARV or UV-ARV induces CD8+ TILs expressing IFN-γ through the TLR3/NF-κB signaling pathway. In this work, CD8+ TILs were treated with the TLR3 inhibitor followed sensitization with ARV or UV-ARV. The results in Fig. 7B-C showed that the TLR3 inhibitor significantly decrease the expression levels of IFN-γ in ARV or UV-ARV-sensitized CD8+ TILs. Having shown that ARV or UV-ARV could induce CD8+ TILs secretion of IFN-γ (Fig. 7B-C), we next wanted to examine whether the upstream signaling of IFN-γ. NF-κB is an inducible transcription factor that is involved in the cytokine-induced immune response [13, 25]. As shown in Fig. 7B-C, treatment with the NF-κB inhibitor resulted in reduced expression of IFN-γ in ARV or UV-ARV sensitized-CD8+TILs, suggesting that UV-ARV or ARV induced CD8+TILs secretion of IFN-γ through the NF-κB signaling pathway. Taken together our results suggested that ARV or UV-ARV-induced IFN-γ secretion of CD8+ TILs is dependent on σC-triggering the TLR3/NF-κB/IFN-γ/TRAIL immunogenic apoptosis pathway.
Fig. 7.
ARV σC and UV-ARV σC activate CD8+ TILs through the TLR3/NF-κb/IFN-γ pathway. ARV- or UV-ARV σC-interacted CD8+ TILs release IFN-γ via the TLR3-dependent NF-κB signaling pathway. (A) Proximity ligation assays for cell-surface TLR3 on CD8+ TILs. The interaction between ARV σC or UV-ARV σC and TLR3 (CD8+ TILs) was assessed by PLA. Representative images are from three independent experiments. Cell nuclei were stained with DAPI (blue). (B, C) Analysis of IFN-γ production by ARV or UV-ARV sensitized-CD8+TILs treated with TLR3 inhibitor or NF-κB inhibitor. CD8+TILs were pretreated with or without TLR3 inhibitor (10 µg/mL) for 30 min and then sensitized with UV-ARV or ARV for 24 h. CD8+TILs were incubated for 1 h with or without 10 µM BAY11-7082 and then sensitized with ARV or UV-ARV for 24 h. IFN-γ and CD8+ was measured by flow cytometry
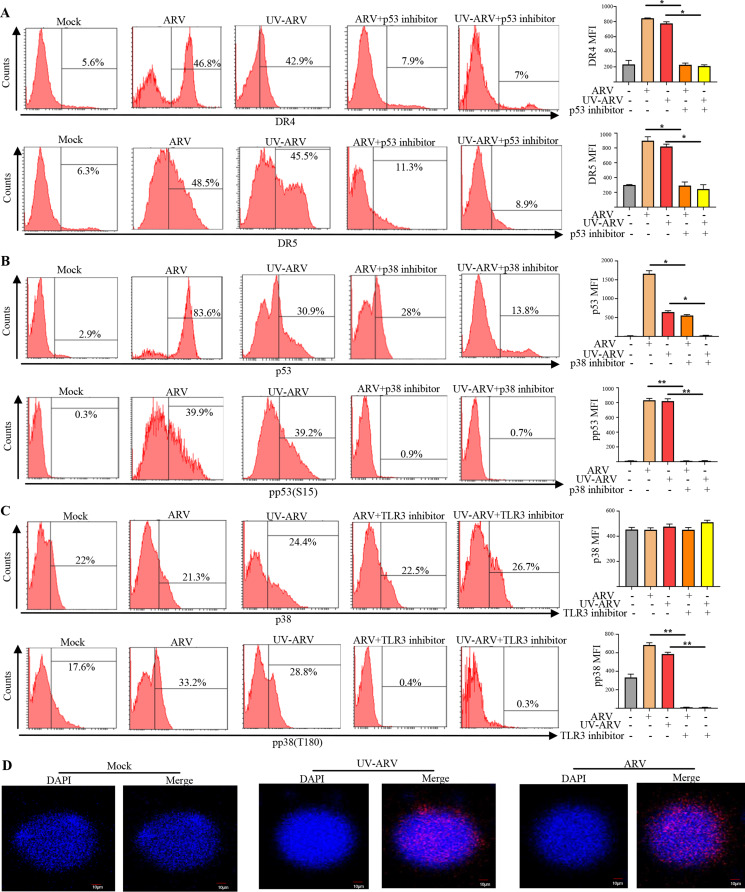
Upregulation of the DR4 and DR5 expression through the p38/p53 signaling pathway in ARV or UV-ARV-sensitized PGC cells
Our results indicated that ARV or UV-ARV treatments upregulates DR4 and DR5 expression on PGC cells by flow cytometry (Fig. 8A). Since DR4 and DR5 are transmembrane domains and cytoplasmic domains of TRAIL receptors [27], which are upregulated on PGC cell surface by ARV or UV-ARV, this directed us to further confirm whether ARV-induced apoptosis of PGC cells occurs due to host signal transduction pathway. Our previous study indicated that σC induces apoptosis in cultured cells and activates a proapoptotic signal by linking p38 to p53 [23]. We next wanted to elucidate whether the p38/p53 signaling pathway upregulates the expression of DR4 and DR5. In this work, PGC cells were pre-treated with the p53 inhibitor for 5 h followed by treatments with ARV or UV-ARV. The results shown in Fig. 8A indicated that the p53 inhibitor significantly decreased the expression levels of DR4 and DR5 in ARV or UV-ARV-sensitized PGC cells. Furthermore, our results revealed that inhibition of p38 by the inhibitor significantly decreased the expression levels of p53 and p-p53 (S15) in ARV or UV-ARV-sensitized PGC cells (Fig. 8B). Treatment with the TLR3 inhibitor reduced the phosphorylated form of p-p38 (T180) in ARV or UV-ARV sensitized-PGC cells, suggesting that UV-ARV or ARV upregulates DR4 and DR5 expression on PGC cells through the p38/p53 signaling pathway (Fig. 8C).
Fig. 8.
The ARV or UV-ARV σC protein interacting with TLR3 of PGC cells and upregulation of DR4 and DR5 death receptors in ARV or UV-ARV-sensitized PGC cells through the p38/p53 signaling pathway. (A) Cell surface staining for DR4 and DR5 of PGC cells from patients was performed in cells pretreated with the p53 inhibitor for 5 h followed by treatment with ARV or UV-ARV. The working concentration for p53 inhibitor was 20 µM. Data are also presented as the ratio between MFI (Median fluorescence intensity) of patients. (B) Intracellular staining for p53 and p-p53(S15) of PGC cells treated ARV or UV-ARV were performed in presence of p38 inhibitor (20 µM). (C) p-p38 (T180) and p38 intracellular staining of PGC cells patients with or without TLR3 inhibitor (10 µg/mL) for 30 min followed by sensitization with UV-ARV or ARV for 24 h. (D) Proximity ligation assays for cell-surface TLR3 on PGC cells. The interaction between ARV σC or UV-ARV σC and TLR3 (PGC cells) was assessed by PLA. Representative images are from three independent experiments. Cell nuclei were stained with DAPI (blue)
Surface expression of TLR3 has been reported in various cancers and TLR3 occur both in the cell membrane and intracellularly, and it seems that activation of the immune response can be initiated concurrently from these two sites in the cell [28]. To study the upstream signaling, we investigated whether ARV or UV-ARV σC protein interacts with the surface TLR3 on PGC cells. Interactions between σC protein with the surface TLR3 of PGC cells were analyzed by in situ PLA. Our results revealed that σC protein interacts with TLR3 of PGC cells (Fig. 8D). In contrast, no signal was observed (Fig. 8D). Our previous study suggested that cell entry of avian reovirus follows a caveolin-1-mediated and dynamin-2-dependent endocytic pathway that requires activation of p38 signaling pathway [29] and cancer cell entry of ARV modulated by σC binging to cell receptors triggers endocytosis [30]. Our results for the first time suggested that ARV or UV-ARV-induced DR4 and DR5 expression of PGC cells is dependent on σC-triggering the TLR3/p38/p53/DR4/DR5 pathway.
Discussion
A perfect OV should eliminate cancer cells through a combination of three mechanisms: including induction of apoptosis, pro-inflammatory cytokines, and IFNs [31, 32]. Besides directly kill tumor cells, OV can activate immune responses or express healing factors to increase antitumor efficacy and enhances efficacy of cancer immune oncological therapy [33, 34]. OV-mediated apoptosis may trigger anticancer immune responses in TEM [35]. Modulation of apoptosis is beginning as a new immunotherapeutic approach for the treatment of cancer [36]. The novel discovery that oncolytic ARV-modulated upregulation of the TLR3/NF-kB/IFN-γ/TRAIL pathway in CD8+TILs, triggering PGC cells of apoptosis through the TLR3/p38/p53/DR4/DR5 pathway.
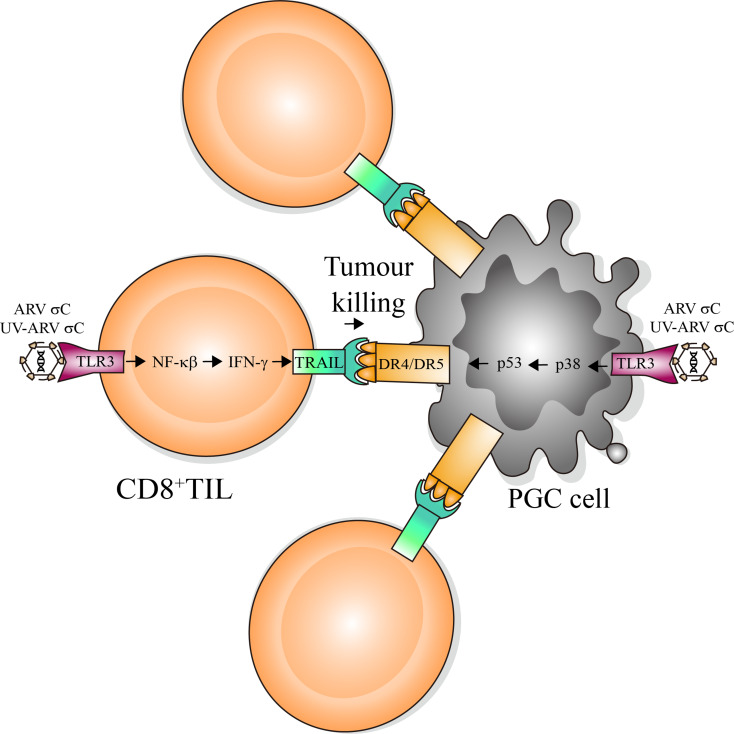
Previous studies suggested that IFN-γ mediates apoptosis of kidney tubular epithelial cells [37] and induces apoptosis through the Jak/Stat pathway by the type I IFN receptor in human colon cancer cells [38]. Although the innate antiviral system of cancer cells may be resistant to the treatment of oncolytic ARV, interestingly, IFN-γ does not inhibit ARV-induced TRAIL expression and ARV-modulated TRAIL-induced apoptosis, suggesting that ARV-induced apoptosis was more sensitive to the TRAIL. Our finding is supported by a previous report suggesting that TRAIL has been implicated in having the IFN-γ response promotor [39]. This study provides a mechanistic insight into ARV- or UV-ARV-sensitized CD8+ TILs expressing TRAIL through activation of the TLR3/NF-kB/IFN-γ pathway preferentially killing PGC cells by immunogenic apoptosis. A model illustrating ARV and UV-ARV-sensitized CD8+ TILs killing PGC cells is outlined in Fig. 9.
Fig. 9.
Schematic diagram showing the ARV or UV-ARV-induced DR4/DR5 expression of PGC cells is dependent on σC-triggering the TLR3/P38/P53/DR4/DR5 pathway. ARV σC and UV-ARV σC activate CD8+TILs to induce immunogenic apoptosis through the TLR3/NF-κb/IFN-γ/TRAIL pathway. ARV σC and UV-ARV σC activate CD8+TILs to kill PGC cells. TILs: Tumor-infiltrating lymphocytes. PGC cells: Primary gastric cancer cells
The use of OV to treat cancer either directly kill OV-infected tumor cells or increase their susceptibility to cell death or apoptosis [40]. A previous study provides potential strategies in cancer treatments with OV and adjuvant NK cells in a cancer treatment [41]. It was reported that TRAIL-armed oncolytic poxvirus suppresses lung cancer cells by inducing apoptosis [42]. Lal et al. developed recombinant measles virus armed with BNiP3 (a pro-apoptotic gene of human origin) as an oncolytic agent to induce apoptosis in breast cancer cells in vitro [43]. Importantly, our findings reveal that oncolytic ARV could be an effective therapeutic strategy for treatment of gastric cancers. This study provides a better insight into in vitro mechanistic immunological studies bridging a systemic model and possibly enable the development of ARV targeted immunomodulatory therapies.
Multiple signaling pathways commonly involved in viral clearance, including IFNs, TLRs, and double-stranded RNA-activating protein kinase (PKR) pathways, may be defective or inhibited in cancer cells, allowing OV to enter and survive in these cells [12, 44]. We have demonstrated for the first time that UV-ARV and ARV σC protein interacts with surface TLR3 of CD8+ TILs and PGC cells. Interestingly, our in situ PLA revealed different staining phenotypes between CD8+ TILs and PGC cells. Oncolytic ARV can exclusively replicate and infect cancer cells (PGC cells), but it is unable to infect healthy cells (CD8+ TILs) [4, 45]. We have demonstrated that ARV infects Vero, DF-1 and AGS cancer lines through σC binding to cellular receptors [30], thereby triggering cavolin and dynamin 2-dependent endocytosis and signaling activation [29, 30). Our study shows that the interaction between the ARV σC protein and the cell surface TLR3 occurs in a ring surrounding CD8+ tumor-infiltrating lymphocytes, while it is evenly distributed on the surface of primary gastric cancer cells (PGC cells).
MRV displays tropism and efficiently replicates in tumor cells with the activated Ras pathway [46]. These characteristics allow the use of MRV in virotherapy, either alone or combined with the conventional and nonconventional treatments [40, 46]. For instance, synergistic cytotoxicity of MRV in combination with cisplatin-paclitaxel doublet chemotherapy [46]. Currently, REOLYSIN®, a formulation of MRV, is used in cancer therapeutics, which has been tested at the preclinical stage and phases I-III clinical studies in a broad range of cancer indications [46]. Our evidences suggest that the antitumoral mechanism associated with ARV or UV-ARV involves the activation of immune response to immunogenic apoptosis. ARV or UV-ARV optimized to attract immune cells to express TRAIL might favorably change the TME. Furthermore, reactive expression of TRAIL in the TME could be a mechanism of resistance to cancer, which induced by IFN-γ. Recent evidence suggests that NK cells can recognize viruses themselves, as in the case of cytomegalovirus which promotes the generation of memory-like NK cells in humans and have an increased IFN-γ and cytolytic response on encounter with target cells [47]. In this study, treatments of PGC cells with ARV and UV-ARV induced a systemic antitumor CD8+ cells response, prominent infiltration of cytotoxic T lymphocytes and Th1 type polarization. Th1 cells produce cytokines, particularly IFN-γ, which play a role in activation and enhancement of cytotoxic T cell expansion and effector functions [45, 48]. Although activated cytotoxic T cells are present in many human tumors, but tumors fail to undergo spontaneous regression [11, 25]. CD8+TILs were found to have an altered phenotype and an impaired ability to secrete IFN-γ. Importantly, oncolytic ARVs or UV-ARVs exert a regulatory role to enhance response of CD8+TILs in the TME. This study demonstrates ARV- or UV-ARV-modulated direct interaction between TILs and PGC cells in an in vitro co-culture system. In our co-culture model, ARV or UV-ARV virotherapy induced a strong CD8+TILs immunity that is therapeutically effective against PGC cells. These support the preclinical development of ARV and UV-ARV as an adjuvant to treat human gastric cancer. This study sheds further light on the molecular basis behind ARV and UV-ARV and facilitates the future efficacy of ARV and UV-ARV as a cancer therapeutic.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
All authors made substantive intellectual contributions to the present study and approved the final manuscript. H.J.L. conceived of the study and generated the original hypothesis, wrote the paper, and supervised the project; Y.Y.W. performed most of the experiments. I.C.C., F.H.W., T.L.L., Y.Y.W., and M. M. analyzed data. I.C.C., F.H.W., T.L.L., Y.Y.W. performed statistical analysis. H.J.L., M.M. revised and edited the manuscript.
Funding
This work was supported by grants from Ministry of Science and Technology of Taiwan (112-2313-B-005-050-MY3), The iEGG and Animal Biotechnology Center from The Feature Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan (112S0023A and 113S0023A), Taichung Veterans General Hospital (TCVGH-NCHU-1127605 and TCVGH-NCHU-1137607).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Patient consent for publication not required.
Ethical approval
The protocol was approved by the ethics committee of Hospital.
Consent for publication
Patient consent for publication not required.
Provenance and peer review
Not commissioned; externally peer reviewed.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fukuhara H, Ino Y, Todo T. Oncolytic virus therapy: a new era of cancer treatment at dawn. Cancer Sci. 2016;107:1373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozak RA, Hattin L, Biondi MJ, Corredor JC, Walsh S, Xue-Zhong M, Manuel J, McGilvray ID, Morgenstern J, Lusty E, Cherepanov V, McBey BA, Leishman D, Feld JJ, Bridle B, Nagy É. Replication and oncolytic activity of an avian orthoreovirus in human hepatocellular carcinoma cells. Viruses. 2017;9:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiu HC, Huang WR, Liao TL, Chi PI, Nielsen BL, Liu JH, Liu HJ. Mechanistic insights into avian reovirus p17-modulated suppression of cell cycle CDK-cyclin complexes and enhancement of p53 and cyclin H interaction. J Biol Chem. 2018;293:12542–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai R, Meng G, Li Y, Wang W, Diao Y, Zhao S, Feng Q, Tang Y. The oncolytic efficacy and safety of avian reovirus and its dynamic distribution in infected mice. Exp Biol Med (Maywood). 2019;244:983–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiu HC, Huang WR, Wang YY, Li JY, Liao TL, Nielsen BL, Liu HJ. Heterogeneous nuclear ribonucleoprotein A1 and lamin A/C modulate nucleocytoplasmic shuttling of avian reovirus p17. J Virol. 2019;93:e00851–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Carli S, Wolf JM, Gräf T, Lehmann FKM, Fonseca ASK, Canal CW, Lunge VR, Ikuta N. Genotypic characterization and molecular evolution of avian reovirus in poultry flocks from Brazil. Avian Pathol. 2020;49:611–20. [DOI] [PubMed] [Google Scholar]
- 7.Huang WR, Li JY, Wu YY, Liao TL, Nielsen BL, Liu HJ. p17-modulated Hsp90/Cdc37 complex governs oncolytic avian reovirus replication by chaperoning p17, which promotes viral protein synthesis and accumulation of viral proteins σC and σA in viral factories. J Virol. 2022;96:e0007422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu HJ, Lin PY, Wang LR, Hsu HY, Liao MH, Shih WL. Activation of small GTPases RhoA and Rac1 is required for avian reovirus p10-induced syncytium formation. Mol Cells. 2008;26:396–403. [PubMed] [Google Scholar]
- 9.Chulu JL, Lee LH, Lee YC, Liao SH, Lin FL, Shih WL, Liu HJ. Apoptosis induction by avian reovirus through p53 and mitochondria-mediated pathway. Biochem Biophys Res Commun. 2007;356:529–35. [DOI] [PubMed] [Google Scholar]
- 10.Lin PY, Liu HJ, Chang CD, Chang CI, Hsu JL, Liao MH, Lee JW, Shih WL. Avian reovirus S1133-induced DNA damage signaling and subsequent apoptosis in cultured cells and in chickens. Arch Virol. 2011;156:1917–29. [DOI] [PubMed] [Google Scholar]
- 11.Boulch M, Cazaux M, Loe-Mie Y, Thibaut R, Corre B, Lemaître F, Grandjean CL, Garcia Z, Bousso P. A cross-talk between CAR T cell subsets and the tumor microenvironment is essential for sustained cytotoxic activity. Sci Immunol. 2021;6:eabd4344. [DOI] [PubMed] [Google Scholar]
- 12.Rommelfanger DM, Compte M, Grau MC, Diaz RM, Ilett E, Alvarez-Vallina L, Thompson JM, Kottke TJ, Melcher A, Vile RG. The efficacy versus toxicity profile of combination virotherapy and TLR immunotherapy highlights the danger of administering TLR agonists to oncolytic virus-treated mice. Mol Ther. 2013;21:348–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furman D, Davis MM. New approaches to understanding the immune response to vaccination and infection. Vaccine. 2015;33:5271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan X, Gajan A, Chu Q, Xiong H, Wu K, Wu GS. Developing TRAIL/TRAIL death receptor-based cancer therapies. Cancer Metastasis Rev. 2018;37:733–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar V, Giacomantonio MA, Gujar S. Role of myeloid cells in oncolytic reovirus-based cancer therapy. Viruses. 2021;13:654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan HF, Kuete M, Su L, Yang F, Hu ZY, Tian BZ, Zhang HP, Zhao K. Comparison of three isolation techniques for human peripheral blood mononuclear cells: cell recovery and viability, population composition, and cell functionality. Biopreserv Biobank. 2016;14:410–5. [DOI] [PubMed] [Google Scholar]
- 17.Aziz F, Yang X, Wen Q, Yan Q. A method for establishing human primary gastric epithelial cell culture from fresh surgical gastric tissues. Mol Med Rep. 2015;12:2939–44. [DOI] [PubMed] [Google Scholar]
- 18.Smoot DT, Sewchand J, Young K, Desbordes BC, Allen CR, Naab T. A method for establishing primary cultures of human gastric epithelial cells. Methods Cell Sci. 2000;22:133–6. [DOI] [PubMed] [Google Scholar]
- 19.Dudley ME, Wunderlich JR, Shelton TE, Even J, Rosenberg SA. Generation of tumor-infiltrating lymphocyte cultures for use in adoptive transfer therapy for melanoma patients. J Immunother. 2003;26:332–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doumba PP, Nikolopoulou M, Gomatos IP, Konstadoulakis MM, Koskinas J. Co-culture of primary human tumor hepatocytes from patients with hepatocellular carcinoma with autologous peripheral blood mononuclear cells: study of their in vitro immunological interactions. BMC Gastroenterol. 2013;13:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vergis J, Malik SVS, Pathak R, Kumar M, Kurkure NV, Barbuddhe SB, Rawool DB. Exploring galleria mellonella larval model to evaluate antibacterial efficacy of cecropin A (1–7)-melittin against multi-drug resistant enteroaggregative Escherichia coli. Pathog Dis. 2021;79:ftab010. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Y, Chen CL, Jiang SW, Feng Y, Yuan L, Chen P, Zhang L, Huang S, Li J, Xia JC, Zheng M. Retrospective analysis of the efficacy of adjuvant CIK cell therapy in epithelial ovarian cancer patients who received postoperative chemotherapy. Oncoimmunology. 2019;8:e1528411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin PY, Lee JW, Liao MH, Hsu HY, Chiu SJ, Liu HJ, Shih WL. Modulation of p53 by mitogen-activated protein kinase pathways and protein kinase C δ during avian reovirus S1133-induced apoptosis. Virology. 2009;385:323–34. [DOI] [PubMed] [Google Scholar]
- 24.Barili V, Fisicaro P, Montanini B, Acerbi G, Filippi A, Forleo G, Romualdi C, Ferracin M, Guerrieri F, Pedrazzi G, Boni C, Rossi M, Vecchi A, Penna A, Zecca A, Mori C, Orlandini A, Negri E, Pesci M, Massari M, Missale G, Levrero M, Ottonello S, Ferrari C. Targeting p53 and histone methyltransferases restores exhausted CD8+ T cells in HCV infection. Nat Commun. 2020;11:604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tabiasco J, Devêvre E, Rufer N, Salaun B, Cerottini JC, Speiser D, Romero P. Human effector CD8 + T lymphocytes express TLR3 as a functional coreceptor. J Immunol. 2006;177:8708–13. [DOI] [PubMed] [Google Scholar]
- 26.Liu WH, Wang X, You N, Tao KS, Wang T, Tang LJ, Dou KF. Efficient enrichment of hepatic cancer stem-like cells from a primary rat HCC model via a density gradient centrifugation-centered method. PLoS ONE. 2012;7:e35720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mérino D, Lalaoui N, Morizot A, Schneider P, Solary E, Micheau O. Differential inhibition of TRAIL-mediated DR5-DISC formation by decoy receptors 1 and 2. Mol Cell Biol. 2006;26:7046–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mielcarska MB, Bossowska-Nowicka M, Toka FN. Cell surface expression of endosomal toll-like receptors-A necessity or a superfluous duplication? Front Immunol. 2021;11:620972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang WR, Wang YC, Chi PI, Wang L, Wang CY, Lin CH, Liu HJ. Cell entry of avian reovirus follows a caveolin-1-mediated and dynamin-2-dependent endocytic pathway that requires activation of p38 mitogen-activated protein kinase (MAPK) and src signaling pathways as well as microtubules and small GTPase rab5 protein. J Biol Chem. 2011;286:30780–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang WR, Wu YY, Liao TL, Nielsen BL, Liu HJ. Cell entry of avian reovirus modulated by cell-surface annexin A2 and adhesion G protein-coupled receptor latrophilin-2 triggers src and p38 MAPK signaling enhancing caveolin-1-and dynamin 2-dependent endocytosis. Microbiol Spectr. 2023;11:e0000923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahmoud MH, Badr G, Badr BM, Kassem AU, Mohamed MS. Elevated IFN-alpha/beta levels in a streptozotocin-induced type I diabetic mouse model promote oxidative stress and mediate depletion of spleen-homing CD8+ T cells by apoptosis through impaired CCL21/CCR7 axis and IL-7/CD127 signaling. Cell Signal. 2015;27:2110–9. [DOI] [PubMed] [Google Scholar]
- 32.Frumence E, Roche M, Krejbich-Trotot P, El-Kalamouni C, Nativel B, Rondeau P, Missé D, Gadea G, Viranaicken W, Desprès P. The South Pacific epidemic strain of Zika virus replicates efficiently in human epithelial A549 cells leading to IFN-β production and apoptosis induction. Virology. 2016;493:217–26. [DOI] [PubMed] [Google Scholar]
- 33.Yang C, Hua N, Xie S, Wu Y, Zhu L, Wang S, Tong X. Oncolytic viruses as a promising therapeutic strategy for hematological malignancies. Biomed Pharmacother. 2021;139:111573. [DOI] [PubMed] [Google Scholar]
- 34.Sobhanimonfared F, Bamdad T, Sadigh ZA, Choobin H. Virus specific tolerance enhanced efficacy of cancer immuno-virotherapy. Microb Pathog. 2020;140:103957. [DOI] [PubMed] [Google Scholar]
- 35.Ma J, Ramachandran M, Jin C, Quijano-Rubio C, Martikainen M, Yu D, Essand M. Characterization of virus-mediated immunogenic cancer cell death and the consequences for oncolytic virus-based immunotherapy of cancer. Cell Death Dis. 2020;11:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lecoultre M, Dutoit V, Walker PR. Phagocytic function of tumor-associated macrophages as a key determinant of tumor progression control: a review. J Immunother Cancer. 2020;8:e001408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du C, Guan Q, Yin Z, Zhong R, Jevnikar AM. IL-2-mediated apoptosis of kidney tubular epithelial cells is regulated by the caspase-8 inhibitor c-FLIP. Kidney Int. 2005;67:1397–409. [DOI] [PubMed] [Google Scholar]
- 38.Sun Y, Qiao L, Xia HH, Lin MC, Zou B, Yuan Y, Zhu S, Gu Q, Cheung TK, Kung HF, Yuen MF, Chan AO, Wong BC. Regulation of XAF1 expression in human colon cancer cell by interferon beta: activation by the transcription regulator STAT1. Cancer Lett. 2008;260:62–71. [DOI] [PubMed] [Google Scholar]
- 39.Häcker S, Dittrich A, Mohr A, Schweitzer T, Rutkowski S, Krauss J, Debatin KM, Fulda S. Histone deacetylase inhibitors cooperate with IFN-gamma to restore caspase-8 expression and overcome TRAIL resistance in cancers with silencing of caspase-8. Oncogene. 2009;28:3097–110. [DOI] [PubMed] [Google Scholar]
- 40.Goldufsky J, Sivendran S, Harcharik S, Pan M, Bernardo S, Stern RH, Friedlander P, Ruby CE, Saenger Y, Kaufman HL. Oncolytic virus therapy for cancer. Oncolytic Virother. 2013;2:31–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aspirin AP, de Los Reyes VAA, Kim Y. Polytherapeutic strategies with oncolytic virus-bortezomib and adjuvant NK cells in cancer treatment. J R Soc Interface. 2021;18:20200669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu J, Wang H, Gu J, Liu X, Zhou X. Trail armed oncolytic poxvirus suppresses lung cancer cell by inducing apoptosis. Acta Biochim Biophys Sin (Shanghai). 2018;50:1018–27. [DOI] [PubMed] [Google Scholar]
- 43.Lal G, Rajala MS. Combination of oncolytic measles virus armed with BniP3, a pro-apoptotic gene and paclitaxel induces breast cancer cell death. Front Oncol. 2018;8:676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heggen-Peay CL, Cheema MA, Ali RA, Schat KA, Qureshi MA. Interactions of poult enteritis and mortality syndrome-associated reovirus with various cell types in vitro. Poult Sci. 2002;81:1661–7. [DOI] [PubMed] [Google Scholar]
- 45.Wu YY, Sun TK, Chen MS, Munir M, Liu HJ. Oncolytic viruses-modulated immunogenic cell death, apoptosis and autophagy linking to virotherapy and cancer immune response. Front Cell Infect Microbiol. 2023;13:1142172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roulstone V, Twigger K, Zaidi S, Pencavel T, Kyula JN, White C, McLaughlin M, Seth R, Karapanagiotou EM, Mansfield D, Coffey M, Nuovo G, Vile RG, Pandha HS, Melcher AA, Harrington KJ. Synergistic cytotoxicity of oncolytic reovirus in combination with cisplatin-paclitaxel doublet chemotherapy. Gene Ther. 2013;20:521–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forrest C, Gomes A, Reeves M, Male V. NK cell memory to cytomegalovirus: implications for vaccine development. Vaccines (Basel). 2020;8:394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim KB, Choi YH, Kim IK, Chung CW, Kim BJ, Park YM, Jung YK. Potentiation of Fas- and TRAIL-mediated apoptosis by IFN-gamma in A549 lung epithelial cells: enhancement of caspase-8 expression through IFN-response element. Cytokine. 2002;20:283–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.