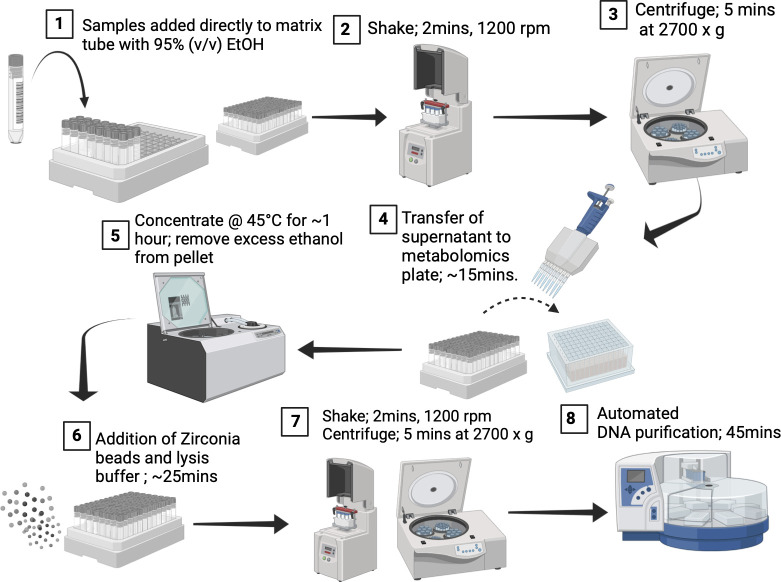
Fig 2.
Flowchart of Matrix method for sample accession, metabolite extraction, and DNA extraction. Step 1: samples are added directly into Matrix Tubes containing 400 µL of 95% (vol/vol) ethanol. Step 2: samples are homogenized using the SpexMiniG plate shaker (SPEXSamplePrep, NJ, USA) at 1,200 rpm for 2 minutes. Step 3: the Matrix rack is centrifuged for 5 min at 2,700 × g. Step 4: Matrix Tubes are de-capped by the automated instrument, Capit-All (Thermo Fisher Scientific, MA, USA). Supernatant is transferred from Matrix Tubes to corresponding wells of a 96-well metabolomics plate (catalog number: 75870-792, VWR), using a multi-channel pipette. Supernatant can later be analyzed by LC-MS/MS. Step 5: Excess ethanol is removed from matrix tubes using a SpeedVac (Thermo Fisher Scientific, MA, USA; catalog number: SPD2030A-220) set to 45°C for 60 min. Step 6: 30 µL of 0.1, 0.5, and 1 mm zirconia-silica beads are added to each matrix tube using the LabTie bead dispenser (MolGen). We add 600 µL of lysis buffer to each Matrix Tube. Matrix Tubes are capped by Capit-All. Step 7: bead beating is performed at 1,200 rpm for 2 min. The Matrix rack is centrifuged for 5 minutes at 2,700 × g. Step 8: Matrix Tubes are de-capped. Lysate is transferred from Matrix Tubes to corresponding wells of a 96-deep well plate using a multi-channel pipette. Nucleic acid purification is performed on the lysate using the KingFisher Flex according to the manufacturer's instructions of the MagMAX Microbiome Ultra Nucleic Acid Isolation Kit (Thermo Fisher, MA, USA). Created with BioRender.com.