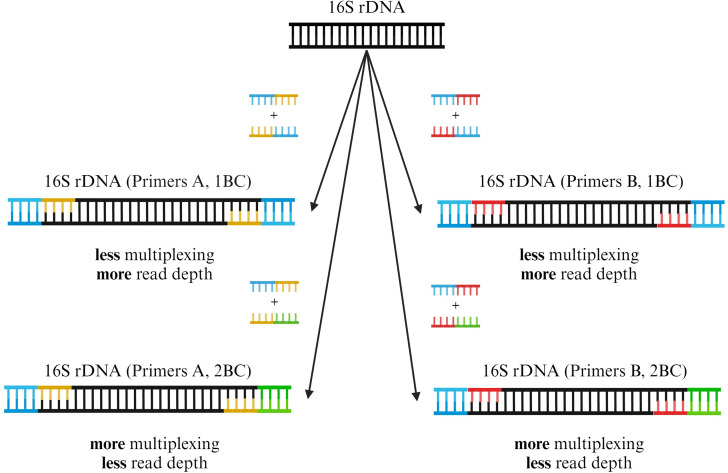
Fig 1.
Barcoded 16S rDNA amplification using different primer pairs and barcodes (1BC and 2BC). Throughout this study, we tested two different barcoded primer pairs for 16S rDNA amplification (A or B). An exemplary 16S rDNA fragment is shown in black. Primer pair A (yellow) is derived from Urban et al. (16). Primer pair B (red) is derived from Matsuo et al. (19) and contains degenerate bases to enhance the amplification of Bifidobacterium spp. in complex microbiological samples. Forward and reverse primers for each primer set were constructed in eight different configurations, each consisting of the primer and one of eight unique barcode sequences. Depending on the desired multiplexing capabilities or sequencing depth, a 1BC (blue) or 2BC (blue and green) approach can be used during PCR preparation with either of the primer pairs A or B. Adding primers with two different barcodes (2BC) allows for a larger sequencing pool but will result in decreased read depth per sample. Utilizing the same barcode in the forward and reverse primers (1BC) results in decreased multiplexing capabilities but enhanced read depth per sample. Created with BioRender.com.