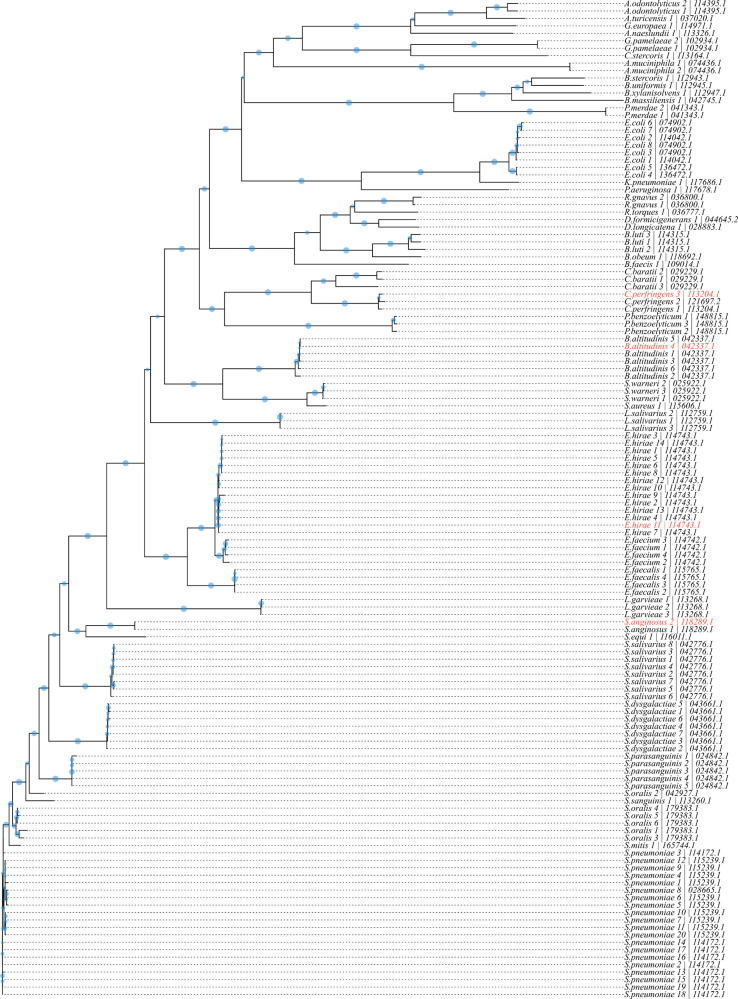
Fig 4.
Phylogenetic tree constructed via the multiple sequence alignment of 134 full-length 16S rDNA consensus sequences in ClustalW2. Branch length is based on the distance matrix generated from pairwise alignment scores. Bootstrapping values (n = 1,000) are indicated as blue circles on the branches (larger circles correspond to larger bootstrapping values). Isolates marked in red were wrongly identified by our sequencing workflow.