Fig. 3.
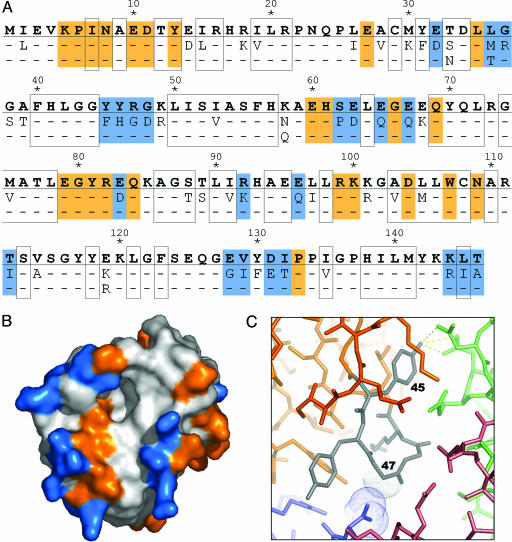
Sequence variation efficiently samples crystal-packing space by modifying the surface properties of the enzyme. (A) The amino acid sequence corresponding to one of the WT gat genes (GenBank accession no. AX543338) is shown in bold, and the 50 positions that vary among at least one of the 11 enzymes tested for crystallization are indicated below. Buried residues are boxed. Residues mediating protein–protein contacts in the crystal are shaded orange (constant positions) or blue (variable positions). (B) Solvent-accessible surface representation of GAT, colored as above. (C) Close-up showing a surface loop (residues 45–49; gray) of one GAT molecule (orange) that mediates protein–protein contacts with three symmetry-related GAT molecules (colored blue, red, and green) in the crystal. The sequence variation at position 45 (Phe vs. Tyr) and 47 (Arg vs. Gly) illustrates two different types of changes that contribute to the formation of a well ordered crystal lattice (see text for details).