Extended Data Fig. 3 |. High-resolution cryo-EM structure of the endogenous core-TIM23 complex.
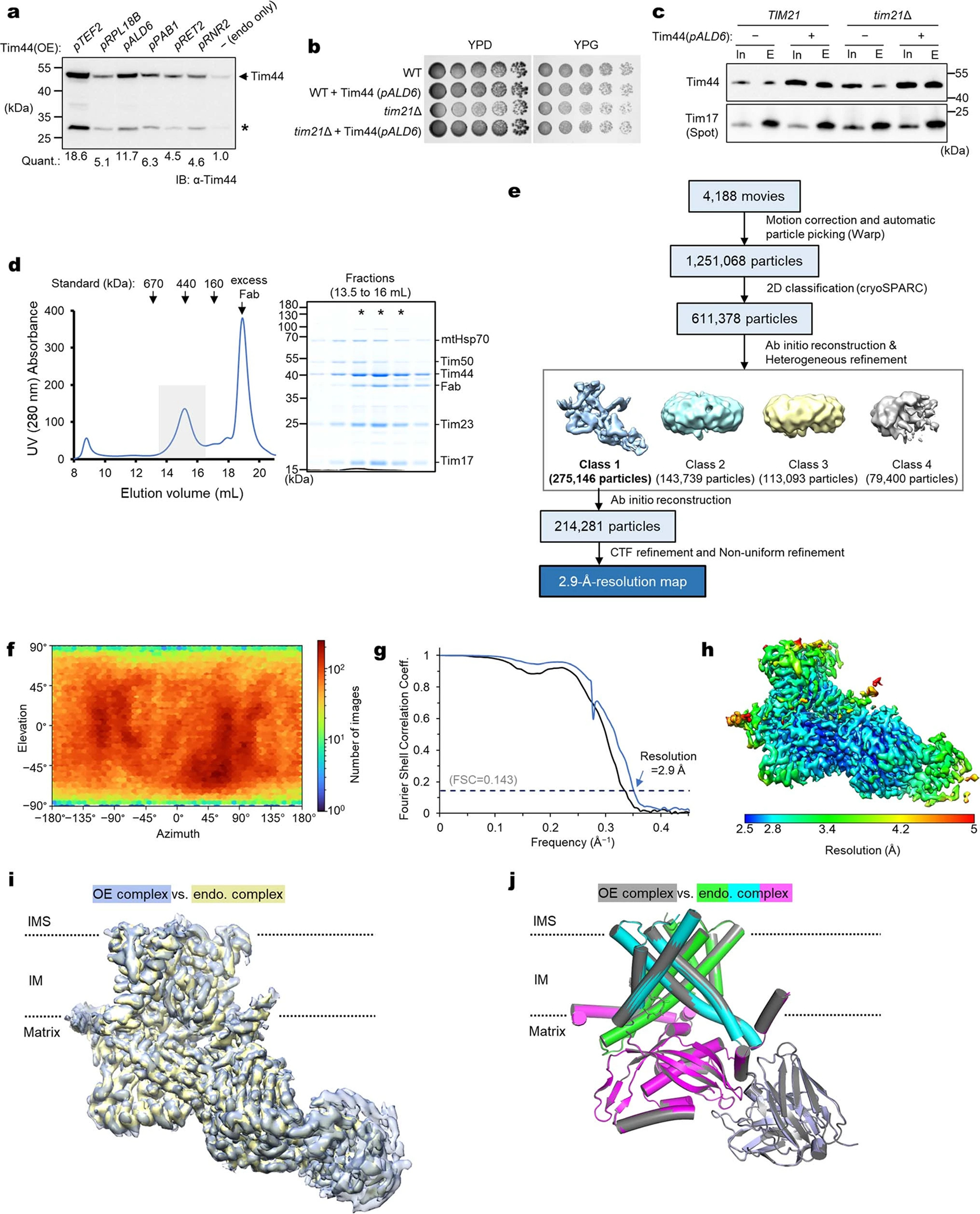
a, To increase Tim44 occupancy in the core TIM23 complex formed by endogenous Tim17 and Tim23, Tim44 was overexpressed under indicated constitutive promoters, and protein levels were analyzed in yeast whole cell lysates (Quants., relative band intensities of full-length Tim44). For the subsequent analyses, we chose the ALD6 promoter (pALD6). Asterisk indicates partial degradation product. b, ALD6-promoter-driven overexpression of Tim44 does not exhibit any cell growth defect. Because the non-essential subunit Tim21 may reduce the amount of the Tim44-containing TIM23 complex by forming a “TIM23-sort” complex12,84, we also tested a chromosomal deletion of tim21. c, Tim17 was immunoprecipitated from the mitochondrial lysate of the strains used in b and co-precipitated Tim44 was detected by immunoblotting (In, input; E, eluate). Note that a substantially increased amount of Tim44 was co-purified with Tim44 overexpression. d, Purification of the Fab-bound, endogenous core-TIM23 complex from the tim21Δ/pALD6-Tim44 strain. Left, Superose-6 SEC elution profile; right, Coomassie-stained SDS gel of fractions indicated in the left panel by a gray box. Fractions marked with asterisks were pooled and used for cryo-EM analysis. Note the substantially increased Tim44:Tim23 ratio compared to the purification shown in Extended Data Fig. 1b. e, Summary of single particle analysis of the endogenous core-TIM23 complex. f–h, Particle view distribution (f), Fourier shell correlation (g), and local resolution distribution (h). i, Superposition of the endogenous core-TIM23 cryo-EM map (blue) onto the TIM23 map (yellow) from overexpression (OE) (Extended Data Fig. 2). The map correlation coefficient was 0.985 in UCSF Chimera. j, As in i, but showing superposition of the atomic models. The RMSD value is 0.26 Å for 739 out of 760 aligned Cα atoms. Data shown in a–d are representative of two experiments.
