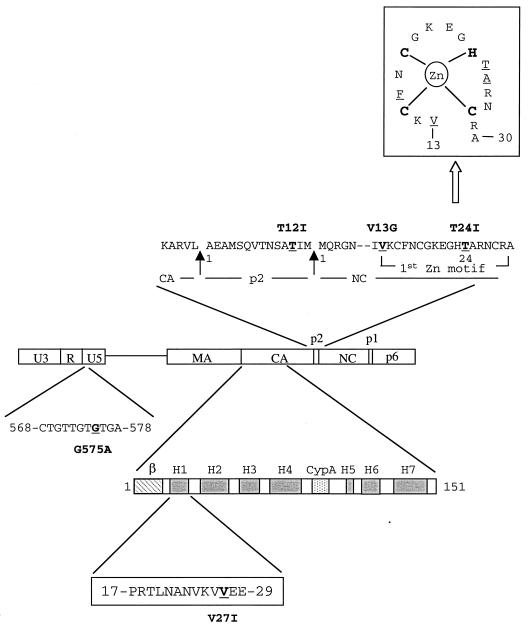
FIG. 3.
Schematic illustration of mutations in the U5 region, CA, p2, and NC. The amino acid sequence of p2, as well as parts of the CA and NC proteins, is shown. The arrows indicate viral protease cleavage sites that result in formation of p2. The substituted nucleotide in U5 or amino residues in CA, p2, and NC are underlined. Nucleotides in U5 are numbered from the beginning of the U3 region. The numbering of the amino acid positions starts from the first residue of each protein. Secondary structural domains in the amino terminus of CA protein (amino acids 1 to 151) are illustrated that include a β-hairpin (β), seven α-helixes (H1 to H7), and a cyclophilin A (Cyp A) binding site located between H4 and H5 (14). The V27I substitution in CA exists in H1. A sketch of the first Zn finger motif, in which some of the second-site mutations are located, is shown at the top of the figure. Interactions between the C, C, H, and C amino residues and the Zn ion are indicated; moreover, the underlined V13, F16, T24, and A25 amino acid residues are close in tertiary structure and form a hydrophobic cleft that can interact with the loop nucleotides of the region of viral genomic RNA SL3 (12).