FIGURE 1.
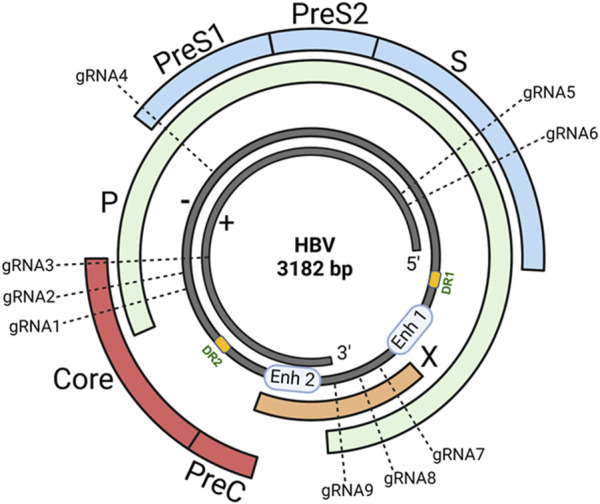
HBV genome and gRNAs design. The genome of HBV is a double-stranded (+/−) circular DNA of about 3.2 kb pairs comprising four overlapping open reading frames (ORFs): surface (S), capsid (C), polymerase (P), and X genes. To select the most specific targets in the HBV genome, we used an online CHOPCHOP program to analyze HBV sequences that have the best-predicted on-target and lowest off-target effects. We compared the genomic DNA sequences of different HBV genotypes originating from distinct geographic regions worldwide and identified 9 target sites in highly conserved regions of the HBV genome (based on the ayw strain, Genbank accession number: NC_003977.2) within the S, C, P, and X genes. The sequence alignments of the gRNA target sites on the HBV genome are shown in our recent publication (Suzuki et al., 2021).
