Abstract
The co-occurrence of multiple chronic conditions, termed multimorbidity, presents an expanding global health challenge, demanding effective diagnostics and treatment strategies. Chronic ailments such as obesity, diabetes, and cardiovascular diseases have been linked to metabolites interacting between the host and microbiota. In this study, we investigated the impact of co-existing conditions on risk estimations for 1375 plasma metabolites in 919 individuals from population-based Estonian Biobank cohort using liquid chromatography mass spectrometry (LC–MS) method. We leveraged annually linked national electronic health records (EHRs) data to delineate comorbidities in incident cases and controls for the 14 common chronic conditions. Among the 254 associations observed across 13 chronic conditions, we primarily identified disease-specific risk factors (92%, 217/235), with most predictors (93%, 219/235) found to be related to the gut microbiome upon cross-referencing recent literature data. Accounting for comorbidities led to a reduction of common metabolite predictors across various conditions. In conclusion, our study underscores the potential of utilizing biobank-linked retrospective and prospective EHRs for the disease-specific profiling of diverse multifactorial chronic conditions.
Keywords: Comorbidities, Metabolomics, Chronic disease, Risk factors, Electronic health records, Biobank
Subject terms: Predictive markers, Risk factors, Microbiome, Cardiovascular diseases, Metabolomics
Introduction
The onset and progression of chronic diseases are influenced by a combination of factors, including genetics, environment, lifestyle, and the microbiome1–3. The etiology of chronic diseases extends beyond isolated conditions, as many chronic conditions share a well-established set of common clinical and lifestyle risk factors4,5. Perturbation of common pathways suggests a significant level of connectivity, and understanding how these associations relate to coexistence of chronic conditions is paramount. One out of three patients suffer from two or more chronic conditions, termed multimorbidity6,7. This is exemplified in individuals with gout, where 74% experience hypertension, and 71% exhibit stage 2 chronic kidney disease8. Therefore, it is essential to distinguish disease-specific biomarkers from those which reflect the progression of other concurrent diseases, termed comorbidities9. For accurate profiling of disease patterns, in-depth health information encompassing each patients’ medical history is needed.
Recent decades have introduced an era of systematic data collection by the establishment of large population-based biobanks, which have been fundamental for the identification of risk factors for chronic diseases10. These biobanks actively recruit participants from the general population and accumulate large sample collections, characterized by comprehensive health data from questionnaires and/or electronic health records (EHR), along with multiple omics data layers. The rich health data facilitates the identification of individuals with diverse disease conditions along with the history of comorbidities and hence becomes indispensable for disentangling risk factors of chronic diseases11. Biobanks also serve as invaluable resources for epidemiological and clinical studies, wherein stored blood samples can be retrospectively analyzed using advanced analytics methods, including high-throughput metabolomics.
Nevertheless, the limited availability of such large cohorts with available follow-up data has resulted in only a few studies aimed at identifying metabolic risk factors across common chronic conditions12–15. While these studies have revealed similarities in association signatures across diseases with distinct pathophysiologies, a comprehensive understanding of disease-specific and shared risk factors remains elusive. Notably, among the mentioned studies, only one has incorporated high-sensitivity mass spectrometry (MS) to determine metabolite profiles for risk estimation12. Compared to nuclear magnetic resonance spectroscopy-derived metabolomic profiles, MS presents a more valuable option as it facilitates the measurement of gut-derived and modulated biomolecules, including bile acids, short-chain fatty acids, branched-chain amino acids, methylamines, tryptophan, and indole derivatives16,17. These biomolecules are modified in metabolic disorders and may serve as significant risk factors for the onset of chronic conditions12,18.
In this study, we investigated the onset of 14 chronic conditions in 991 Estonian Biobank (EstBB) participants and their associations with baseline levels of 1,375 plasma metabolites measured with untargeted MS profiling. The primary aim of this study was to identify and distinguish disease-specific and common metabolites which are contributing to the risk of chronic diseases. For this, we used the EHR information to uncover comorbidity profiles of all cases and specific disease-naive controls selected from a population cohort.
Results
Study overview and data description
We studied a well-phenotyped cohort of 991 individuals from the Estonian Biobank with available untargeted plasma metabolite data generated by Metabolon HD4 platform. Following the exclusion of individuals with missing data (see Methods), the analysis comprised 919 participants (63.1% females), with an average follow-up time of 11.0 years (SD 4.4 years). To evaluate the effect of the metabolite levels on the risk of developing chronic diseases, 14 common conditions with more than 40 incident cases were evaluated (Fig. 1, Supplementary Table S1). The mean age and body mass index at sampling was 46.7 years (SD 16.8) and 26.8 kg/m2 (SD 5.7), respectively. Other characteristics of the study population are listed in Supplementary Table S2.
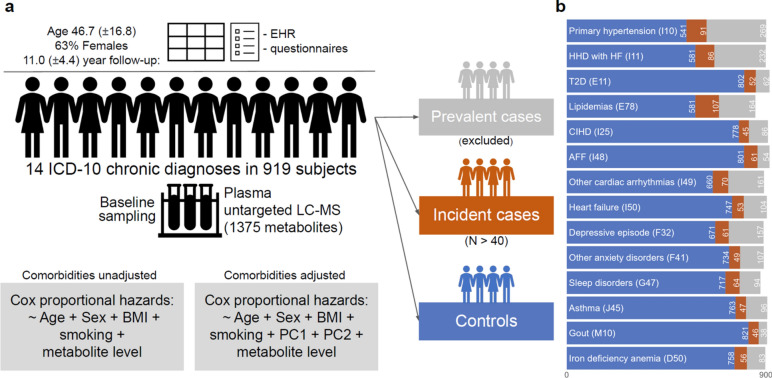
Fig. 1.
Design of the study. a Analysis plan. b Counts of controls (blue, never diagnosed with the respective condition), incident cases (red, first diagnosed with the respective condition after sample collection), prevalent cases (grey, first diagnosed with the respective condition before sample collection, excluded from further analysis) for selected diseases. 14 chronic conditions with more than 40 incident cases were studied. Cox proportional hazard models were adjusted for age, sex, bmi, smoking status in the primary analysis. In the secondary analysis, Cox models were additionally adjusted by the first two principal components (PC) of Hamming distance between comorbidity presence/absence profiles of the study subjects. AFF—atrial fibrillation and flutter, HHD with HF—hypertensive heart disease with heart failure, CIHD—chronic ischemic heart disease, T2D—type 2 diabetes, LC–MS—liquid chromatography mass spectrometry.
Metabolomics analysis was performed on plasma samples collected between 2002 and 2018. Subsequently, following quality checks and the exclusion of infrequent and drug-related metabolites (see Methods), association analysis was conducted on 1375 metabolites (Supplementary Table S3). To evaluate the impact of comorbidities on risk estimates, we performed a secondary analysis by adjusting for comorbidities (Fig. 1a). Additionally, all accessible metabolites were cross-referenced with recent literature data to determine their association with the gut microbiome.
Plasma metabolites predict the onset of various chronic conditions
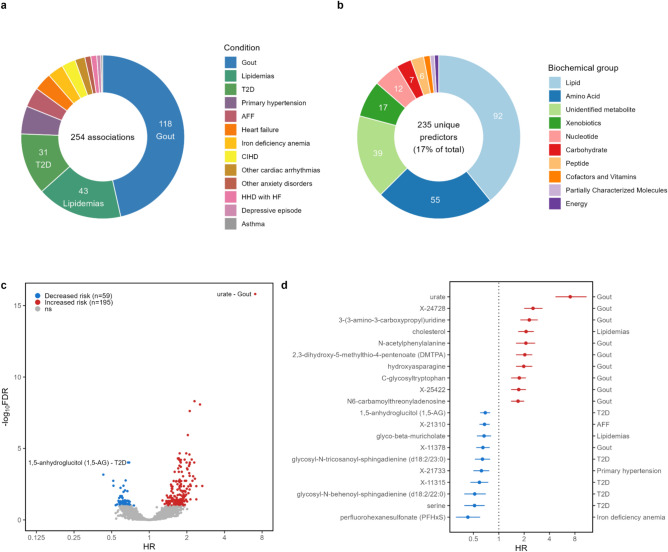
To investigate the role of plasma metabolites in the incidence of 14 chronic conditions, Cox proportional hazards models adjusted for age, sex, body mass index (BMI), and smoking status were constructed. In total, we detected 254 significant associations (false discovery rate, FDR < 0.1) with 13 incident diseases (Fig. 2a, Supplementary Fig. S1). Notably, for sleep disorders, we did not find any significant associations. A total of 17% (235/1375) metabolites were significantly (FDR < 0.1) linked to at least one disease (Fig. 2b, Supplementary Fig. S1, Supplementary Table S4). The largest proportion of the associated metabolites were linked with risk of developing gout (n = 118). A substantial number of metabolites showed an association with lipidemias (n = 43) and type 2 diabetes (T2D, n = 31). At the same time, primary hypertension (n = 14) and several cardiac conditions showed a lower number of associations: atrial fibrillation and flutter (AFF, n = 10), heart failure (n = 9), chronic ischemic heart disease (CIHD, n = 7), other cardiac arrhythmias (n = 5), hypertensive heart disease with heart failure (HHD with HF, n = 3). Similarly, only a few associations were detected for iron deficiency anemia (n = 8), other anxiety disorders (n = 3), depressive episode (n = 2), and asthma (n = 1). The significant predictors predominantly comprised lipids (n = 92), amino acids (n = 55), and unidentified metabolites (n = 39) (Fig. 2b, Supplementary Table S4 online). Consistent with previous studies, we observed significant associations between uric acid and the increased risk of gout (hazard ratio, HR 7), as well as cholesterol and the increased risk of lipidemias (HR 2.1) (Fig. 2d). Elevated uric acid levels, indicative of hyperuricemia, are a known risk factor of gout, suggesting that both metabolites may mirror metabolic changes in the pre-disease state19.
Fig. 2.
Associations between plasma metabolome and risk of 14 chronic diseases using random controls (FDR < 0.1). a Total number of significant associations with incident diseases. b Total number of significant predictors divided into biochemical groups. c Volcano plot of the hazard ratios (HR) and FDR values of incident risk factors for chronic conditions. d Top 10 associations with both increased and decreased risk of incident diseases. AFF—atrial fibrillation and flutter, HHD with HF—hypertensive heart disease with heart failure, CIHD—chronic ischemic heart disease, T2D—type 2 diabetes. Cox models were adjusted for age, body mass index, sex, and smoking status. Error bars show the 95% confidence interval.
The five strongest associations with each incident condition are depicted in Supplementary Fig. S2 online. Notably, among the best predictors of gout and lipidemias, strong correlations were observed within incident cases of respective conditions (Supplementary Fig. S2, Supplementary Table S5 online). On the contrary, the top predictors for T2D exhibited relatively low levels of correlation indicating potentially higher heterogeneity among these metabolites and related pathways.
The majority of identified interactions indicated an increased risk (Fig. 2c, Supplementary Fig. S1 online). Among the top ten associations with highest HR, nine were specifically associated with the development of incident gout (Fig. 2d). In contrast, when examining associations with negative HR indicating diminished risk with higher metabolite values, the situation was more variable—five out of ten of the most significant predictors were identified in relation to incident T2D.
Chronic conditions have partially overlapping metabolic predictors
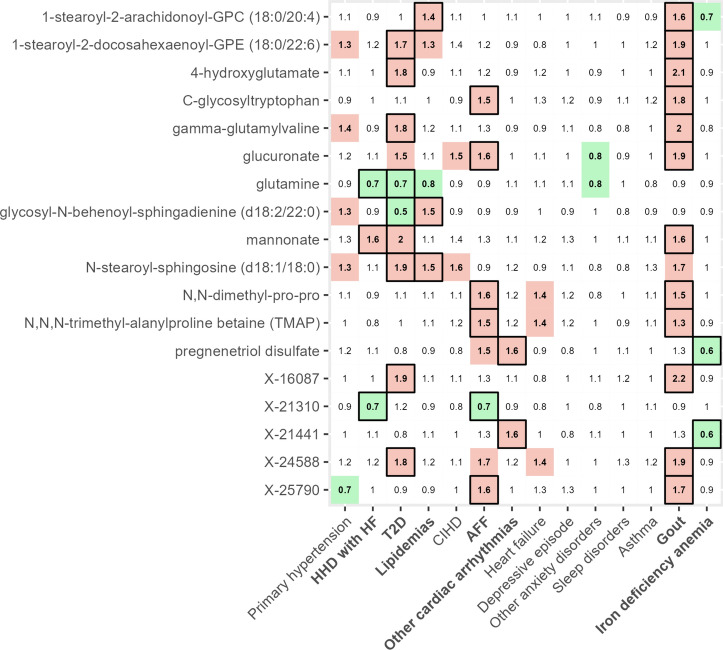
We investigated the extent of unique and shared metabolic predictors for diseases. Most of the reported predictors (92%, 217/235) were uniquely associated with the risk of a single disease (Suppl Fig. S3 online). Shared associations were detected mainly between two chronic diseases (17 predictors) and no common predictors were seen between more than 3 diseases (Fig. 3).The highest number of shared associations appeared between gout and T2D (n = 6), gout and AFF (n = 5), and gout and lipidemias (n = 3). For example, higher level of mannonate was associated with increased risk of incident gout (HR 1.6), T2D (HR 2), and HHD with HF (HR 1.6), corroborating a previous study, which demonstrated association of higher mannonate levels with severe insulin-deficiency20. An unidentified metabolite X-24588 was associated with incident gout (HR 1.9) and T2D (HR 1.8). This metabolite has been previously associated with hepatic triglyceride content21.
Fig. 3.
Common risk factors of chronic conditions. Heatmap illustrates hazard ratios of metabolites shared between at least two conditions (FDR < 0.1 significant values are encased in black frames; green highlight—nominally significant reduced risk; red highlight—nominally significant increased risk). AFF—atrial fibrillation and flutter, HHD with HF—hypertensive heart disease with heart failure, CIHD—chronic ischemic heart disease, T2D—type 2 diabetes. Cox models were adjusted for age, body mass index, sex, and smoking status.
Disease-specific associations with chronic conditions are more robust to adjusting for comorbidities
While comorbidities are commonly overlooked during control selection, we next aimed to integrate them into our analysis. To achieve this, we employed principal component analysis based on Hamming distance between comorbidity presence/absence profiles of the study subjects. In the secondary analysis, we additionally adjusted Cox proportional hazards models by the first two principal components. This adjustment aimed to address the differences in the disease burden between incident cases and controls, thereby strengthening the reliability of our findings. For instance, 56% of incident cases of AFF in the EstBB cohort were already diagnosed with concurrent primary hypertension, whereas only 22% of randomly selected participants without incident AFF exhibited primary hypertension at the time of sampling.
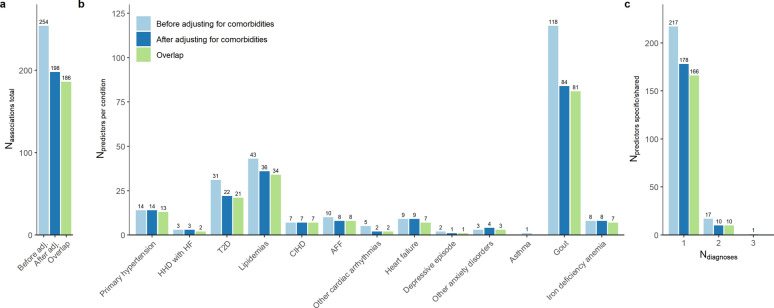
In the sensitivity analysis, a lower total number of significant associations (198 vs 254, 73% overlap) and unique predictors (188 vs 235, 75% overlap) were observed compared to analysis conducted without adjusting for comorbidities (Fig. 4a, Supplementary Fig. S4 and S5, Supplementary Table S4 online). Reduction in the number of associations was more evident in the case of gout (84 vs 118), lipidemias (36 vs 43), and T2D (22 vs 31) (Fig. 4b). The incident cases for these diseases were inflated by comorbid prevalent diagnoses, potentially explaining the loss of signal in the secondary analysis (Supplementary Table S6 online).
Fig. 4.
Comparison of results from primary and comorbidity-adjusted analyses. A comparison of results of primary analysis (before adjusting for comorbidities, black) and secondary analysis (after adjusting for comorbidities, dark gray). Light gray highlights the overlap of significant associations/predictors between the two approaches. a Total number of associations. b Number of predictors across evaluated conditions. c Number of condition-specific (Ndiagnoses = 1) and shared predictors (Ndiagnoses > 1). AFF—atrial fibrillation and flutter, HHD with HF—hypertensive heart disease with heart failure, CIHD—chronic ischemic heart disease, T2D—type 2 diabetes. Cox models for primary analysis were adjusted for age, body mass index, sex, smoking status. Cox models for sensitivity analysis were further adjusted by the first two principal components calculated from Hamming distances between comorbidity profiles.
Overall, employing comorbidities as covariates led to reduction in both the total number of disease-specific (178 vs 217) and shared risk predictors (10 vs 18) (Fig. 4c). The more pronounced decrease among common predictors could suggest that these associations were confounded by the presence of comorbid conditions (Supplementary Fig. S6, Supplementary Table S6 online).
In total, 166 disease-specific associations were consistently identified, resulting in a 76% overlap with previously identified metabolites from the primary analysis. This suggests the existence of a more robust set of predictors exclusively associated with each specific medical condition, as opposed to a limited number of widespread incident risk signals shared among multiple chronic conditions.
A substantial portion of risk factors for chronic conditions are linked with microbiota
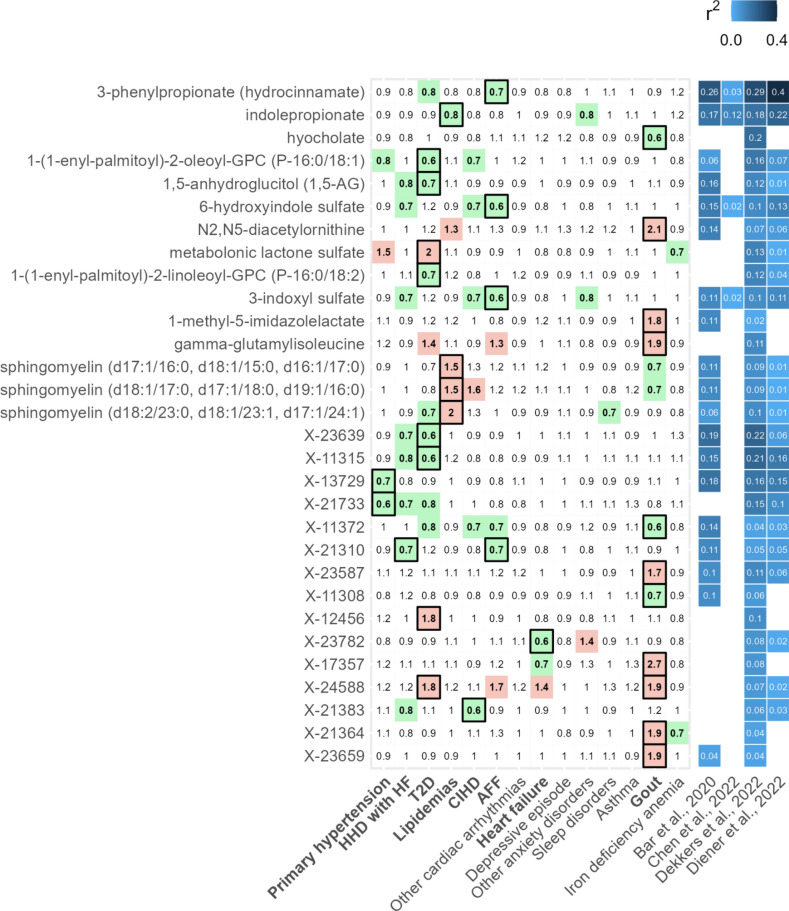
The growing evidence of the microbial activity on metabolites prompted us to pay attention to disease-associated metabolites with pre-established significant associations to the gut microbiome. We extracted and aggregated the data from four recent publications which reported microbiota-explained variance of individual serum or plasma metabolites22–25. This analysis revealed notable microbiome contributions for 93% (219/235) of significant and 78% (892/1140) of non-significant predictors (see Supplementary Table S4 online). Within the prominent microbiome-associated metabolites (with any reported R2 value exceeding 0.1) significant associations were shown for T2D, lipidemias, primary hypertension, HHD with HF, AFF, and gout (Fig. 5, Supplementary Fig. S7 online). These associations were predominantly exclusive to a single disease and included a high number of unidentified metabolites. Notably, 3-phenylpropionate (hydrocinnamate) and hyocholate were associated with reduced risk of AFF and gout, respectively, while well-established cardiometabolic markers 1,5-anhydroglucitol (1,5-AG) and metabolonic lactone sulfate were linked to decreased and increased risk of incident T2D, respectively26–28. Among microbial metabolites originating from amino acids, indolepropionate displayed a reduced risk of incident lipidemias, while 3-indoxyl sulfate and 6-hydroxyindole sulfate demonstrated a lower risk of incident AFF. However, no associations were found between trimethylamine N-oxide (TMAO), phenylacetylglutamine, or cinnamoylglycine and disease risk, contrasting previous findings29–32. Further discussion is available in the Supplementary Discussion section.
Fig. 5.
Top microbiome-related incident risk factors of chronic conditions. Left—heatmap illustrates hazard ratio values of the 15 foremost microbiome-related identified and unidentified metabolites with at least 1 significant association (FDR < 0.1 significant values are encased in black frames, nominally significant with green or red highlights). Right—heatmap shows metabolite variance explained by the gut microbiota from the analyzed literature22–25. AFF—atrial fibrillation and flutter, HHD with HF—hypertensive heart disease with heart failure, CIHD—chronic ischemic heart disease, T2D—type 2 diabetes. Cox models were adjusted for age, body mass index, sex, and smoking status.
Discussion
In this study, we conducted an untargeted metabolomics analysis of plasma to identify both disease-specific and shared risk factors across 14 chronic conditions in the EstBB subcohort of 991 individuals. We leveraged well-phenotyped population-based biobank data to uncover predictive metabolic markers, demonstrating the utility of this approach. Notably, our findings highlight predominantly disease-specific signals, challenging the notion of widespread commonality among chronic diseases. However, we identified significant shared risk factors for gout, T2D, AFF, and lipidemias, indicating potential metabolic interactions among these conditions. A novel aspect of our study is the observed decrease in shared predictors when adjusting for prevalent comorbidities, underscoring the influence of comorbid conditions on metabolic risk profiles. Additionally, we showed that a high proportion of identified predictors were previously associated with gut microbial composition, suggesting a link between metabolism and the gut microbiome. Importantly, our findings imply that comorbidities may contribute to the shared incident risk signature observed across chronic conditions, offering new insights into the interplay between metabolic markers, gut microbiota, and comorbidities.
The highest number of incident metabolic risk associations was identified for gout, potentially due to its high comorbidity rate and its role as a risk factor for other conditions. Gout is an arthritic condition induced by hyperuricemia leading to urate deposits in the tissues. Previous studies on gout have shown associations with metabolic syndrome and chronic kidney disease33,34. We are not aware of any untargeted metabolomics studies investigating risk factors of gout. Nevertheless, previous research on prevalent gout has established connections with altered amino acid levels, perturbations in purine, glycerophospholipid, sphingolipid, and carbohydrate metabolism35. These findings were also partially replicated in our study. Our study identified over a dozen N-acetylated metabolites exclusively associated with an increased risk of gout, with the strongest signal from N-acetylalanine. These metabolites have also been previously linked to various chronic conditions and impaired kidney function12,36.
Within the studied conditions, we observed a limited (8%) concurrence of metabolite incident risk factors. In contrast, Pietzner et al.12, reported a 65.5% overlap for metabolite predictors among 27 noncommunicable diseases, including, ten cancer types when data was sourced from hospitalization and cancer registry data. This represents a crucial distinction from our study, as we not only obtained data from the aforementioned registries but also integrated EHR data from primary care and other relevant registries. For example, MacRae et al.36, suggested using EHRs from various registries for classification of clinical data as they reported higher age of onset of multimorbidity within the identical patient cohort when relying on information derived from hospitalizations compared to data obtained from primary care sources. This suggests that relying solely on hospitalization data might result in inaccurate estimation of comorbidities, likely influencing findings of disease risks and reported interconnectivity among chronic diseases.
We noticed that, for most of the studied conditions, the prevalence of comorbidities was substantially higher in incident cases compared to controls. In response, we aimed to enhance the analysis by including baseline comorbidities information as additional covariates. This aligns with a recent study emphasizing the need for distinguishing disease-specific changes from confounders from pre- and comorbidities9. More specifically, Fromentin et al. employed a design that incorporated not only healthy and clinically ill individuals but also subjects with dysmetabolic morbidities, enabling the comparison of metabolic signatures across various disease states and clinical stages. Similarly, our approach aimed to disentangle condition-specific effects from the multimorbidity signal. Adjustment for comorbidities resulted in a reduction in both disease-specific and shared predictors, with the most pronounced impact observed in conditions that initially exhibited the highest number of associations, namely, gout, lipidemias, and T2D. Therefore, associations initially thought to be common might be attributed to the presence of shared comorbid conditions rather than being independent associations across various diagnoses. For future studies, a more thorough consideration of distinct comorbidity profiles could enhance the detection of risk factors37. We also propose that utilizing registry-based electronic health record (EHR) data could potentially be expanded to specifically select subjects at various stages of disease progression, each with their respective comorbidity profiles, and corresponding selection of appropriate controls.
We also demonstrated predominantly disease-specific associations among metabolites linked to the microbiome in the external studies we reviewed. For example, indolepropionate and 3-phenylpropionate were exclusively associated with reduced risk of lipidemias and AFF, respectively. Both metabolites have been associated with reduced chronic disease risk12. Also, in the same study by Pietzner et al., levels of these metabolites were not significantly mediated by any of the available routine clinical parameters, including renal markers. Dekkers et al.25 showed that several Eubacteriales sp. and more specifically, Faecalibacterium prausnitzii species are positively associated with aforementioned metabolites25. In addition, multiple associations of microbially produced indole-derived metabolites and reduced risk of chronic diseases were observed. Contrastingly, previous studies have linked indoxyl sulfate with further progression of chronic kidney disease and cardiovascular disease38. Therefore, it could be hypothesized that in individuals with normal renal function, maintaining optimal levels of uremic toxins could protect against cardiovascular issues. Further discussion of the identified metabolites is available in the Supplementary Discussion section.
Our study is set apart from prior research on the simultaneous investigation of metabolite incident disease risk factors by multiple aspects. First, utilization of extensive registry data distinguishes our approach from studies that depend on self-reported or single registry-based data, enhancing the robustness of our findings, and by providing less biased and more objective/standardized diagnosis status of multiple chronic conditions. Second, the inclusion of a wide range of frequently occurring chronic disorders, from cardiac and metabolic conditions to mood disorders, contributes to a comprehensive exploration of risk factors, identifying significant associations for all investigated conditions. Third, alongside the conventional analysis, we adjust risk predictions for comorbidities. This results in a more nuanced evaluation of risk factors specific to diseases, as well as those shared among them. Notably, the potential confounding effects arising from comorbidities might not have been comprehensively addressed in previous studies12. Last, we demonstrate a high number of microbially-associated metabolites among the significant incident disease predictors. This was achieved by integrating recently published data on the explained variance of metabolite levels attributed to the gut microbiome. Through this approach, we were able to link previously established microbiome-metabolite associations to a large proportion of the significant predictors.
This investigation has certain limitations that warrant consideration. Crucially, our study encounters constraints in terms of statistical power due to small sample size in the condition-specific incident case groups. Moreover, the absence of a validation cohort could affect the generalizability of our findings. However, the specific selection of diverse conditions would require extensive collaboration with partner institutions, potentially limited by the wide scope of this study. In addition, we opted not to employ any additional inclusion or exclusion criteria specific to any particular disease. While a single ICD-10-specific definition of chronic conditions might impose limitations, it does provide a standardized and plain approach that facilitates ease of assessment and replication for broad selection of diseases. Finally, we did not account for the confounding effects of treatments, dietary habits, or medication intake.
Conclusion
In conclusion, our study presents a unique contribution to the field by utilizing extensive registry data for exploration of a diverse array of chronic diseases. While acknowledging certain limitations, our research provides valuable insights into understanding the microbial connection and specificity of metabolic predictors for incident chronic diseases. Our findings highlight the need for further research to consider comorbidities as additional confounders in assessing disease risk.
Methods
Sample description
In this study, we utilized retrospective and prospective data from well-characterized individuals in the Estonian Biobank (EstBB). Established in 2000, the EstBB encompasses over 210,000 adults (aged 18–93) across Estonia and maintains updated electronic health records (EHR) through regular linkages to primary care, hospital databases, and national registries, including the Cancer Registry and Causes of Death Registry, in addition to the national health insurance fund11,39. Disease and condition records were coded using International Classification of Diseases, 10th revision (ICD-10).
The study cohort comprised 991 individuals who joined the EstBB between 2002 and 2019. During recruitment, participants provided venous blood samples and completed extensive questionnaires covering health-related topics such as lifestyle, diet, and pre-existing ICD-10 coded clinical diagnoses11. 919 individuals with complete covariate information (age, sex, BMI, smoking status) were included in the subsequent analyses.
Chronic conditions were identified and aggregated based on the first three characters of ICD-10 codes from EHR data, enabling the tracking of participants’ health over time and the analysis of both prevalent and incident diseases. Prevalent diseases refer to ongoing or existing conditions that participants had at the time of sample collection. Incident diseases, on the other hand, are new cases of disease that developed during the follow-up period among participants who did not have the condition at the time of sample collection. For this study, we selected 14 chronic diseases with a minimum of 40 incident cases (Fig. 1b, Supplementary Table 1). The date of disease incidence for individuals without a prior diagnosis was defined as the first recorded diagnosis after their enrollment in EstBB. To increase the reliability of diagnosis data, individuals were classified as cases if they had at least two separate diagnosis entries on different dates during the follow-up period. Controls for each target disease were selected by excluding individuals with either incident or prevalent disease status, as well as those with only one diagnosis entry during the follow-up period. Within 919 cases included in the study, 37% had 0 comorbidities, 63% had 1 or more comorbidities. Supplementary Table S6 provides data on incident and prevalent comorbidities within study groups.
Metabolomics profiling
Untargeted metabolomics profiling on EDTA plasma samples stored in -80C was conducted in 2021 using an ultra-performance liquid chromatography coupled to tandem mass spectrometry (UPLC-MS/MS) system (HD4, Metabolon Inc., Durham, USA)40. Raw data were subjected to Metabolon´s standard quality control and processing, including imputation and batch-normalization of peak area data. Specifically, peaks were quantified using the area under the curve. For normalization, peak area measurements for each metabolite were adjusted by dividing by the median peak area of samples within each instrument batch (144 samples per plate). Measurements below the detection threshold were imputed with the minimum observed value for that metabolite. Subsequently, the data were log-transformed, mean-centered, and divided by the standard deviation for further analysis.
Compound identification was performed by comparing the data to Metabolon’s library of standards and recurrent unknown entities. This library contains comprehensive details such as retention time/index, mass-to-charge ratio (m/z), and chromatographic data, including MS/MS spectral data. Metabolites were classified into three tiers based on the level of confirmation: (1) no asterisk: compound confirmed with an authentic chemical standard, (2) single asterisk: compound identified with confidence but not confirmed with a standard, (3) double asterisk: compound reasonably identified without an available standard. Unnamed/unknown metabolites, denoted with the prefix “X-,” represent a yet unidentifed group of reported compounds, consistently detected based on their molecular signature, including retention time, accurate mass, and fragmentation pattern. Additionally, metabolites were annotated into two classification levels: (1) “SUPER_PATHWAY”: broad metabolic pathway categories, and (2) “SUB_PATHWAY”: more specific metabolic pathway categories. Metabolites with fewer than 10 measurements or designated as drugs (SUB_PATHWAY contains “Drug”) were eliminated. Metabolite levels were not adjusted for any treatment, medication or dietary intake data. The employed metabolomics pipeline encompasses well-established gut microbiota-derived metabolites, such as choline metabolites, tryptophan metabolites in kynurenine and indole pathways, and bile acids18. Metadata for all analyzed metabolites can be found in Supplementary Table S3.
Statistical analysis
We employed time-dependent Cox proportional hazards regression models to evaluate the hazard ratio (HR) for incidence events relative to the control group. Cox models were fitted individually for each condition sub-cohort using the R package survival (v3.5.0). All models were adjusted for age, sex, BMI, and by current/previous smoking status. To address any potential imbalance in comorbidity burden between cases and controls, we conducted a secondary analysis supported by Principal Component Analysis (PCA). Briefly, pairwise Hamming distances were calculated from binarized comorbid disease status information for each selected disease and then subjected to PCA. Subsequently, all models were further adjusted by incorporating the first two principal components representing the comorbidity load. P values were corrected for multiple testing by application of Benjamini-Hochberg (B-H) procedure, with a significance threshold set at FDR < 0.1. To investigate the relationships between metabolite levels, the Pearson correlation coefficient was calculated for each pair of metabolites using the R package Hmisc (v5.1.2). This analysis was conducted separately within incident cases for each condition. All statistical analyses were performed and figures created using R v4.1.1
Literature analysis for metabolite and gut microbiome associations
In this study, we conducted a comprehensive analysis of metabolites, integrating data from existing literature to provide an additional layer of information on microbiome-metabolite associations. It’s essential to clarify that the microbiome composition analysis for the respective cases was beyond the scope of this study. We focused on large-scale studies (N > 300) that utilized MS metabolomics and metagenomic sequencing to explore the microbial capacity to predict serum or plasma metabolite levels22–25. Three of these studies (excluding Chen et al., 2022) utilized an untargeted MS platform by Metabolon. Specifically, we extracted explained variance data of metabolites from the supplementary data of the aforementioned publications. Additionally, when reported, we considered interactions with FDR < 0.05 as significant microbiome associations, resulting in 1132 significant associations out of the 1375 studied metabolites. To establish links with metabolites from these studies, we first used Metabolon ID, and when unavailable, we employed Metabolon Chemical Name, and finally Human Metabolome Database (HMDB) ID. For consistency, all HMDB IDs were transformed to 7-digit format, as required, from their original 5- and 6-digit accession numbers.
Supplementary Information
Acknowledgements
The authors would like to thank Mari-Liis Tammesoo, Marili Palover, Neeme Tõnisson, Liis Leitsalu, and Esta Pintsaar for participating in the sample collection process of the EstBB cohort. We also thank Krista Fischer for contributing to the experimental design of this study and acknowledge the Estonian Biobank research team members Andres Metspalu, Tõnu Esko, Mari Nelis, Georgi Hudjashov, and Lili Milani. EstBB Metabolon assays used in this study were funded by Biomarin Pharmaceutical. This work was written at writing retreats and writing days organized by the Institute of Genomics, University of Tartu.
Author contributions
M.J., O.A., E.O., J.K. formulated overall objectives and study design. T.N., U.V., T.E. organized the collection and analysis of the samples. M.J. organized the phenotype and health data from questionnaires and electronic health records. M.J. performed the data analysis. M.J. interpreted the data and prepared the figures. M.J. wrote the first version of the paper. E.O., O.A., A.R., J.K, L.B., K.E., A.W. contributed to the revision of the paper. Estonian Biobank research team collected and provided EstBB data. All authors read and approved the final version of the paper.
Funding
Current work was funded by Estonian Research Council grants (PRG1414 to E.O.) and an EMBO Installation grant (No. 3573 to E.O.). The work of J.K, U.V. and T.E. was supported by the Estonian Research Council grant PRG1291. Part of the analysis was performed on the HPC servers of the University of Tartu.
Data availability
The datasets generated and analyzed during the current study contain sensitive information from healthcare registers and are therefore not publicly available. However, they can be obtained from the corresponding author upon reasonable request. The procedure for accessing the data from the Estonian Biobank is available at https://genomics.ut.ee/en/content/estonian-biobank. The datasets generated and analyzed during the current study are not publicly available since the data access to the Estonian Biobank must follow the informed consent regulations of the Estonian Committee on Bioethics and Human Research, which are clearly described in the Data Access section at https://genomics.ut.ee/en/content/estonian-biobank. Rights of Estonian Biobank’s participants are regulated by Human Genes Research Act (HGRA) § 9 – Voluntary nature of gene donation (https://www.riigiteataja.ee/en/eli/ ee/531,102,013,003/consolide/current). All data access to the Estonian Biobank’s data must adhere to the informed consent regulations established by the Estonian Committee on Bioethics and Human Research. To initiate a request for phenotype data, it is necessary to submit a preliminary request to releases@ut.ee.
Declarations
Competing interests
During the drafting of the manuscript, L.B. is an employee of BioMarin.
Ethics statement and Consent for publication
Estonian biobank conducts all data collection and research activities according to the Estonian Human Genes Research Act (HGRA). Ethical approval was obtained from the Estonian Committee on Bioethics and Human Research (Estonian Ministry of Social Affairs; approval No. 1.1–12/624) and for the data release from EstBB (T06 6–7/GI/8175). Subjects signed a informed consent form during recruitment, and to ensure privacy protection, no personally identifiable information was used in the analyses.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper
Contributor Information
Elin Org, Email: elin.org@ut.ee.
Estonian Biobank research team:
Andres Metspalu, Lili Milani, Tõnu Esko, Reedik Mägi, Mari Nelis, and Georgi Hudjashov
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-75556-1.
References
- 1.Stanaway, J. D. et al. Global, regional, and national comparative risk assessment of 84 behavioural, environmental and occupational, and metabolic risks or clusters of risks for 195 countries and territories, 1990–2017: a systematic analysis for the global burden of disease stu. Lancet392, 1923–1994. 10.1016/S0140-6736(18)32225-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mars, N. et al. Polygenic and clinical risk scores and their impact on age at onset and prediction of cardiometabolic diseases and common cancers. Nat. Med.26, 549–557. 10.1038/S41591-020-0800-0 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Vijay, A. & Valdes, A. M. Role of the gut microbiome in chronic diseases: a narrative review. Eur. J. Clin. Nutr.76, 489–501. 10.1038/S41430-021-00991-6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Calderón-Larrañaga, A. et al. Multimorbidity and functional impairment-bidirectional interplay, synergistic effects and common pathways. J. Intern. Med.285, 255–271. 10.1111/JOIM.12843 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peters, R. et al. Common risk factors for major noncommunicable disease, a systematic overview of reviews and commentary: the implied potential for targeted risk reduction. Ther. Adv. Chronic. Dis.10.1177/2040622319880392 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen, H. et al. Prevalence of multimorbidity in community settings: a systematic review and meta-analysis of observational studies. J. Comorb.10.1177/2235042X19870934 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jürisson, M. et al. Prevalence of chronic conditions and multimorbidity in estonia: a population-based cross-sectional study. BMJ Open. 10.1136/BMJOPEN-2021-049045 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu, Y., Pandya, B. J. & Choi, H. K. Comorbidities of gout and hyperuricemia in the us general population: NHANES 2007–2008. Am. J. Med.10.1016/J.AMJMED.2011.09.033 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Fromentin, S. et al. Microbiome and metabolome features of the cardiometabolic disease spectrum. Nat. Med.28, 303–314. 10.1038/s41591-022-01688-4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinkorová, J. & Topolčan, O. Biobanks in the era of big data: objectives, challenges, perspectives, and innovations for predictive, preventive, and personalised medicine. EPMA J.11, 333. 10.1007/S13167-020-00213-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leitsalu, L. et al. Cohort Profile: Estonian Biobank of the Estonian Genome Center, University of Tartu. Int. J. Epidemiol.44, 1137–1147. 10.1093/IJE/DYT268 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Pietzner, M. et al. Plasma metabolites to profile pathways in noncommunicable disease multimorbidity. Nat. Med.27, 471–479. 10.1038/s41591-021-01266-0 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buergel, T. et al. Metabolomic profiles predict individual multidisease outcomes. Nat. Med.10.1038/s41591-022-01980-3 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Julkunen, H. et al. Atlas of plasma NMR biomarkers for health and disease in 118,461 individuals from the UK Biobank. Nat. Commun.10.1038/S41467-023-36231-7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrett, J. C. et al. Metabolomic and genomic prediction of common diseases in 477,706 participants in three national biobanks. medRxiv. 10.1101/2023.06.09.23291213 (2023).37732231 [Google Scholar]
- 16.Emwas, A. H. et al. NMR spectroscopy for metabolomics research. Metabolites. 10.3390/METABO9070123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou, L. et al. Gut microbiota-related metabolome analysis based on chromatography-mass spectrometry. Trends Anal. Chem.143, 116375. 10.1016/J.TRAC.2021.116375 (2021). [Google Scholar]
- 18.Agus, A., Clément, K. & Sokol, H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut70, 1174. 10.1136/GUTJNL-2020-323071 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, L., Zhang, Y. & Zeng, C. Update on the epidemiology, genetics, and therapeutic options of hyperuricemia. Am. J. Transl. Res.12, 3167 (2020). [PMC free article] [PubMed] [Google Scholar]
- 20.Zaghlool, S. B. et al. Metabolic and proteomic signatures of type 2 diabetes subtypes in an arab population. Nat. Commun.13, 1–17. 10.1038/s41467-022-34754-z (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faquih, T. O. et al. Hepatic triglyceride content is intricately associated with numerous metabolites and biochemical pathways. Liver Int.43, 1458–1472. 10.1111/LIV.15575 (2023). [DOI] [PubMed] [Google Scholar]
- 22.Bar, N. et al. A reference map of potential determinants for the human serum metabolome. Nature588, 135–140. 10.1038/s41586-020-2896-2 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Diener, C. et al. Genome-microbiome interplay provides insight into the determinants of the human blood metabolome. Nat. Metab.4, 1560. 10.1038/S42255-022-00670-1 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen, L. et al. Influence of the microbiome, diet and genetics on inter-individual variation in the human plasma metabolome. Nat. Med.10.1038/s41591-022-02014-8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dekkers, K. F. et al. An online atlas of human plasma metabolite signatures of gut microbiome composition. Nat. Commun.13, 5370. 10.1038/s41467-022-33050-0 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ying, L. et al. Serum 1,5-anhydroglucitol when used with fasting plasma glucose improves the efficiency of diabetes screening in a chinese population. Sci. Rep.7, 1–8. 10.1038/s41598-017-12210-z (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gall, W. E. et al. α-hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PLoS One5, e10883. 10.1371/JOURNAL.PONE.0010883 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das, S. K. et al. Metabolomic architecture of obesity implicates metabolonic lactone sulfate in cardiometabolic disease. Mol. Metab.10.1016/J.MOLMET.2021.101342 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi, J. et al. Circulating trimethylamine N-oxide and the risk of cardiovascular diseases: a systematic review and meta-analysis of 11 prospective cohort studies. J. Cell. Mol. Med.22, 185–194. 10.1111/JCMM.13307 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romano, K. A. et al. Gut microbiota-generated phenylacetylglutamine and heart failure. Circ. Heart Fail.16, E009972. 10.1161/CIRCHEARTFAILURE.122.009972 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu, Y. et al. Two distinct gut microbial pathways contribute to meta-organismal production of phenylacetylglutamine with links to cardiovascular disease. Cell. Host Microbe31, 18-32.e9. 10.1016/j.chom.2022.11.015 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menni, C. et al. Serum metabolites reflecting gut microbiome alpha diversity predict type 2 diabetes. Gut Microbes11, 1632–1642. 10.1080/19490976.2020.1778261 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen, Y. Y. et al. The association of uric acid with the risk of metabolic syndrome, arterial hypertension or diabetes in young subjects- an observational study. Clin. Chim. Acta478, 68–73. 10.1016/J.CCA.2017.12.038 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Kielstein, J. T., Pontremoli, R. & Burnier, M. Management of hyperuricemia in patients with chronic kidney disease: a focus on renal protection. Curr. Hypertens. Rep.10.1007/S11906-020-01116-3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, X. & You, C. The biomarkers discovery of hyperuricemia and gout: proteomics and metabolomics. PeerJ.10.7717/PEERJ.14554 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacRae, C. et al. Impact of data source choice on multimorbidity measurement: a comparison study of 2.3 million individuals in the welsh national health service. BMC Med.21, 1–12. 10.1186/S12916-023-02970-Z/FIGURES/3 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koskinen, M. et al. Data-driven comorbidity analysis of 100 common disorders reveals patient subgroups with differing mortality risks and laboratory correlates. Sci. Rep.10.1038/S41598-022-23090-3 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim, Y. J., Sidor, N. A., Tonial, N. C., Che, A. & Urquhart, B. L. Uremic toxins in the progression of chronic kidney disease and cardiovascular disease: mechanisms and therapeutic targets. Toxins (Basel). 10.3390/TOXINS13020142 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leitsalu, L., Alavere, H., Tammesoo, M. L., Leego, E. & Metspalu, A. Linking a population biobank with national health registries—the Estonian experience. J. Pers. Med.5, 96. 10.3390/JPM5020096 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans, A. M. High resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. J. Postgenomics Drug Biomarker Dev.10.4172/2153-0769.1000132 (2014). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study contain sensitive information from healthcare registers and are therefore not publicly available. However, they can be obtained from the corresponding author upon reasonable request. The procedure for accessing the data from the Estonian Biobank is available at https://genomics.ut.ee/en/content/estonian-biobank. The datasets generated and analyzed during the current study are not publicly available since the data access to the Estonian Biobank must follow the informed consent regulations of the Estonian Committee on Bioethics and Human Research, which are clearly described in the Data Access section at https://genomics.ut.ee/en/content/estonian-biobank. Rights of Estonian Biobank’s participants are regulated by Human Genes Research Act (HGRA) § 9 – Voluntary nature of gene donation (https://www.riigiteataja.ee/en/eli/ ee/531,102,013,003/consolide/current). All data access to the Estonian Biobank’s data must adhere to the informed consent regulations established by the Estonian Committee on Bioethics and Human Research. To initiate a request for phenotype data, it is necessary to submit a preliminary request to releases@ut.ee.